Home >Technology peripherals >AI >Tsinghua AIR and others proposed ESM-AA, the first protein language model from amino acids to atomic scales
Tsinghua AIR and others proposed ESM-AA, the first protein language model from amino acids to atomic scales
- WBOYWBOYWBOYWBOYWBOYWBOYWBOYWBOYWBOYWBOYWBOYWBOYWBOriginal
- 2024-06-28 18:10:061374browse

Research teams from Tsinghua University AIR, Peking University, and Nanjing University proposed the ESM-AA model. This model has made important progress in the field of protein language modeling, providing a unified modeling solution that integrates multi-scale information.
It is the first protein pre-trained language model that can handle both amino acid information and atomic information. The excellent performance of the model demonstrates the great potential of multi-scale unified modeling to overcome existing limitations and unlock new capabilities.
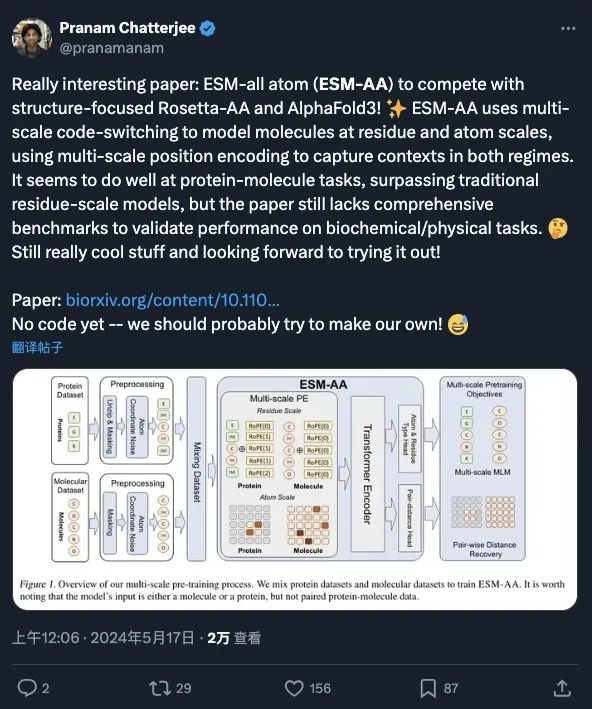
As a base model, ESM-AA has received attention and extensive discussion from many scholars (see screenshot below), and is considered to have the potential to develop a model based on ESM-AA that can compete with AlphaFold3 and RoseTTAFold All-Atom. It opens new avenues to study the interactions between different biological structures. The current paper has been accepted by ICML 2024.
Research background
Protein is the key executor of various life activities. In-depth understanding of proteins and their interactions with other biological structures is a core issue in biological sciences, which has significant practical significance for targeted drug screening, enzyme engineering and other fields.
Therefore, how to better understand and model proteins has become a research hotspot in the field of AI4Science.
In recent days, major cutting-edge research institutions, including Deepmind and the Baker Group of the University of Washington, have also conducted in-depth research on the problem of protein all-atom modeling, and proposed methods including AlphaFold 3, RoseTTAFold All-Atom, etc. for proteins and other life activities. The all-atomic scale modeling model of related molecules can achieve accurate predictions of protein structure, molecular structure, receptor-ligand structure and other all-atomic scale with high accuracy.
Although these models have made significant progress in structural modeling at the all-atomic scale, current mainstream protein language models are still unable to achieve protein understanding and representation learning at the all-atomic scale.
Multi-scale, the "necessary path" for the next generation of protein models
Protein representation learning models represented by ESM-2 use amino acids as the only scale for building models, which is suitable for situations that focus on processing proteins is a reasonable approach.
However, the key to fully understanding the nature of proteins lies in elucidating their interactions with other biological structures such as small molecules, DNA, RNA, etc.
Facing this demand, it is necessary to describe the complex interactions between different structures, and it is difficult for a single-scale modeling strategy to provide effective comprehensive coverage.
To overcome this shortcoming, protein models are undergoing a profound innovation toward multi-scale models. For example, the RoseTTAFold All-Atom model published in Science magazine in early May, as a follow-up product of RoseTTAFold, introduced the concept of multi-scale.
This model is not limited to protein structure prediction, but also extends to broader research fields such as docking of proteins and molecules/nucleic acids, protein post-translational modifications, etc.
At the same time, DeepMind’s newly released AlphaFold3 also adopts a multi-scale modeling strategy to support the prediction of the structure of multiple protein complexes. Its performance is impressive and will undoubtedly have a major impact on the fields of artificial intelligence and biology.
ESM All-Atom, a multi-scale protein language model base
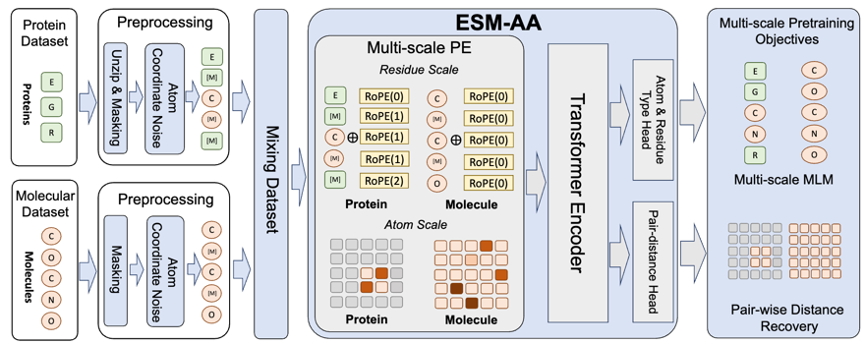
The successful application of RoseTTAFold All-Atom and AlphaFold3 to the multi-scale concept inspired a Important thinking is: how the protein language model as a protein base model should adopt multi-scale technology. Based on this, the team proposed the multi-scale protein language model ESM All-Atom (ESM-AA).
Briefly, ESM-AA introduces the concept of multi-scale by "unzip" some amino acids into the corresponding atomic composition. Subsequently, pre-training was performed by mixing protein data and molecular data, which gave the model the ability to simultaneously handle biological structures of different scales.
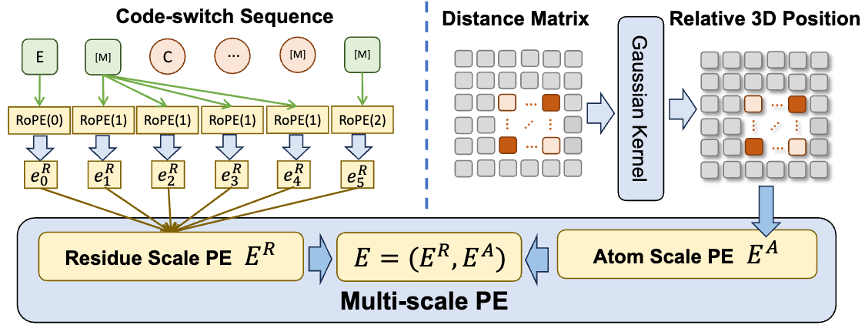
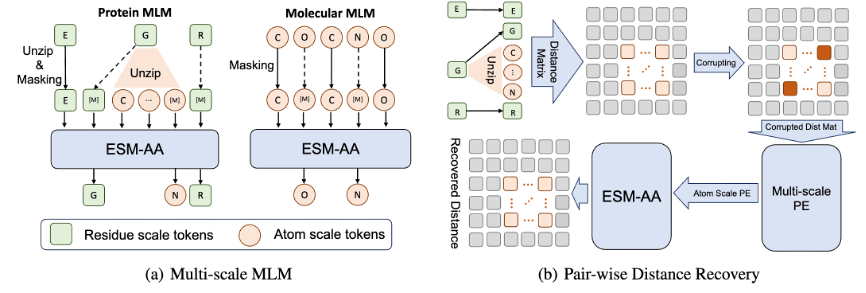
In addition, in order to help the model better learn high-quality atomic scale information, ESM-AA will also use atomic scale molecular structure data for training. Moreover, by introducing the multi-scale position encoding mechanism shown in Figure 2, the ESM-AA model can well distinguish information at different scales, ensuring that the model can accurately understand position and structural information at the residue level and atomic level.
Multi-scale pre-training objectives
To help the model learn multi-scale information, the team designed a variety of pre-training objectives for the ESM-AA model. The multi-scale pre-training objectives of ESM-AA include masked language modeling (MLM) and pairwise distance recovery (PDR). As shown in Figure 3(a), MLM requires the model to make predictions based on the surrounding context by masking amino acids and atoms. This training task can be performed at both the amino acid and atomic scales. PDR requires the model to accurately predict the Euclidean distance between different atoms to train the model to understand atomic-level structural information (as shown in Figure 3(b)).
Experimental verification
Performance evaluation
ESM-AA model is fine-tuned and evaluated on multiple protein-small molecule benchmark tasks, including enzyme-substrate substance affinity regression task (results are shown in Figure 4), enzyme-substrate pair classification task (results are shown in Figure 4), and drug-target affinity regression task (results are shown in Figure 5).
The results show that ESM-AA outperforms previous models in these tasks, indicating that it fully realizes the potential of protein pre-trained language models at the amino acid and atomic scales.
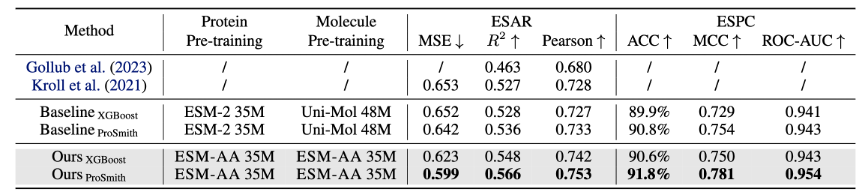
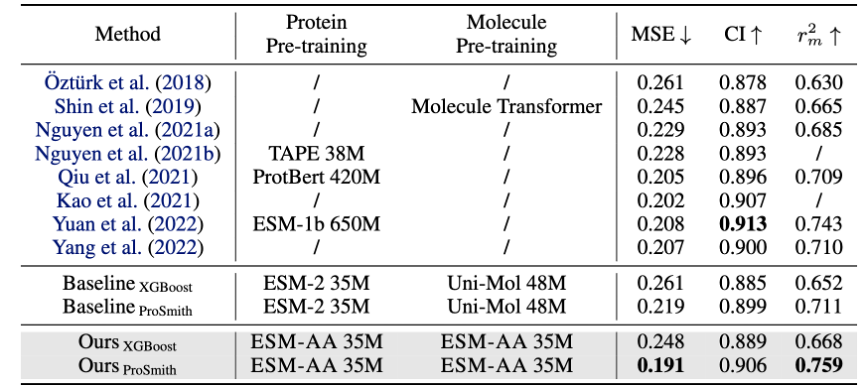
Figure 5: Performance comparison of drug-target affinity regression task
In addition, the ESM-AA model also The performance was tested on tasks such as protein contact prediction, protein functional classification, and molecular property prediction.
The results show that when dealing with tasks involving only proteins, ESM-AA performs on par with ESM-2; on molecular tasks, the ESM-AA model outperforms most benchmark models and is similar to Uni-Mol. .
This shows that ESM-AA does not sacrifice its ability to understand proteins in the process of acquiring powerful molecular knowledge. It also further illustrates that the ESM-AA model successfully reuses the knowledge of the ESM-2 model without having to start from scratch. development, significantly reducing model training costs.
Visual Analysis
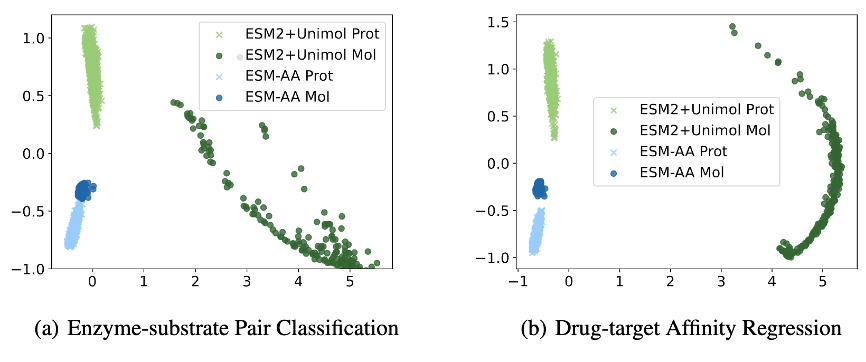
In order to further analyze the reasons why ESM-AA performs well on the protein-small molecule benchmark task, this paper shows the extraction of the ESM-AA model and the ESM-2+Uni-Mol model combination in this task Visualization of sample representation distribution.
As shown in Figure 6, the protein and small molecule representations learned by the ESM-AA model are more compact, which indicates that the two are in the same representation space. This is why the ESM-AA model is better than the ESM-2+Uni- Mol model, further illustrating the advantages of multiscale unified molecular modeling.
Conclusion
The ESM-AA developed by the Tsinghua AIR team is the first protein pre-trained language model that integrates amino acid and atomic information processing. The model demonstrates robust and excellent performance by integrating multi-scale information, providing a new way to solve the problem of interactions between biological structures.
ESM-AA not only promotes a deeper understanding of proteins, but also performs well in multiple biomolecule tasks, proving that it can effectively integrate molecular-level knowledge while maintaining protein understanding capabilities, reducing the cost of model training. It opens up a new direction for AI-assisted biological research.
Paper title: ESM All-Atom: Multi-Scale Protein Language Model for Unified Molecular Modeling
The above is the detailed content of Tsinghua AIR and others proposed ESM-AA, the first protein language model from amino acids to atomic scales. For more information, please follow other related articles on the PHP Chinese website!

