Home >Technology peripherals >AI >Meta creates the first 'Protein Universe' panorama! Using a 15 billion parameter language model, more than 600 million protein structures were predicted
Meta creates the first 'Protein Universe' panorama! Using a 15 billion parameter language model, more than 600 million protein structures were predicted
- PHPzforward
- 2023-04-12 18:25:101495browse
Meta has taken another step forward in the exploration of protein structure!
This time they are targeting a larger target area: metagenomics.
The "Dark Matter" of the Protein Universe
According to the NIH Human Genome Research Institute, metagenomics (Metagenomics, also translated) Metagenomics) the study of the structure and function of entire nucleotide sequences isolated and analyzed from all organisms (usually microorganisms) in a bulk sample, often used to study specific microbial communities such as those living on human skin, soil Proteins from microorganisms in water or water samples.
Over the past few decades, metagenomics has been a very active field as we learn more about all the microorganisms that live in, on, and in humans and in the environment.
Because the research objects of metagenomics are all-encompassing, far exceeding the proteins that make up animal and plant life, it can be said to be the least understood protein on the earth.
To this end, Meta AI uses the latest large-scale language models, creates a database of more than 600 million metagenomic structures, and provides an API that allows scientists to easily search for specific protein structures relevant to their work.
Paper address: https://www.biorxiv.org/content/10.1101/2022.07.20.500902v2
Meta represents, decoding metagenomic structure, It helps to unravel the long-standing mysteries of human evolutionary history, and helps humans cure diseases and purify the environment more effectively.
Protein structure prediction, 60 times faster!
Metagenomics is the study of how to obtain DNA from all these organisms that coexist in the environment. It's a bit like a box of puzzles, but it's not just a box of puzzles, it's actually All 10 sets of smaller puzzles are stacked together and placed in a box.
When metagenomics obtains the genomes of these 10 organisms at the same time, it is actually trying to solve 10 puzzles at the same time and understand all the different puzzle pieces in the same genome box.
It is precisely because of this unknown structure and biological role that new proteins discovered through metagenomics can even be called the "dark matter" of the protein universe.

In recent years, advances in genetic sequencing have made it possible to catalog billions of metagenomic protein sequences.
However, although the existence of these protein sequences is known, it is a huge challenge to further understand their biological properties.
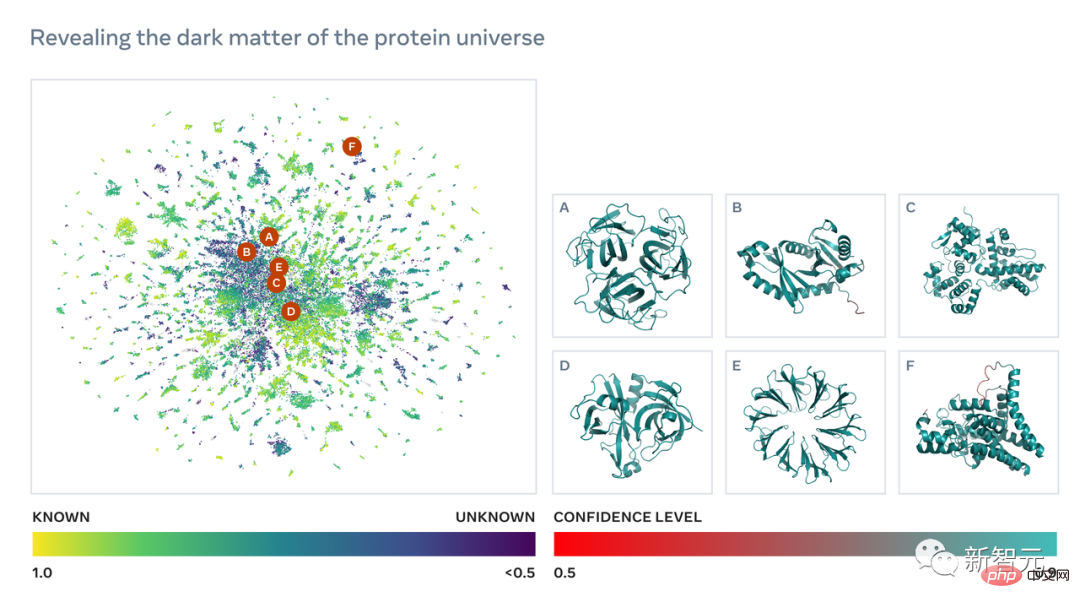
In order to obtain the sequence structures of these billions of proteins, a breakthrough in prediction speed is crucial.
This process, even with the most advanced tools and the computing resources of a large research institution, may take several years.
As a result, Meta trained a large language model to learn evolutionary patterns and generate accurate structure predictions end-to-end directly from protein sequences, while maintaining accuracy and predicting faster than the current state-of-the-art. method is 60 times faster.
In fact, with the help of this new structure prediction capability, Meta predicted more than 600 million metagenomic proteins in the map in just two weeks using a cluster of approximately 2,000 GPUs. the sequence of.
The metagenomic map released by Meta is called ESM Atlas, which almost covers the predictions of the entire metagenomic sequence public database MGnify90.
Meta stated that ESM Atlas is the largest high-resolution predicted structure database to date, 3 times larger than existing protein structure databases, and the first database to comprehensively and large-scale cover metagenomic proteins.
These protein structures provide an unprecedented view into the breadth and diversity of nature and have the potential to accelerate the discovery of proteins with practical applications in areas such as medicine, green chemistry, environmental applications and renewable energy.
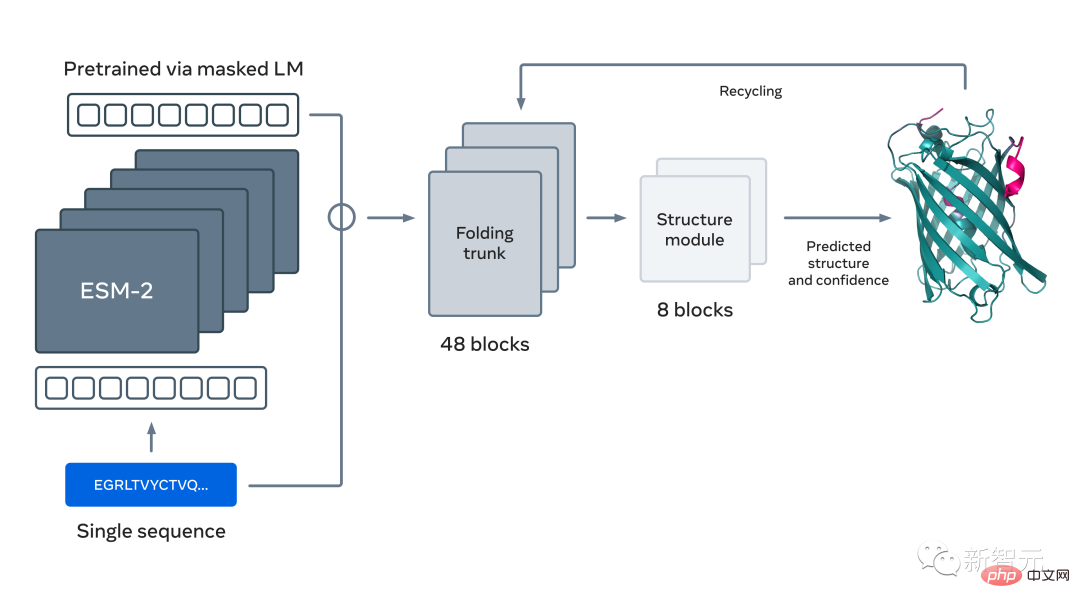
The new language model used to predict protein structure has 15 billion parameters, making it the largest "protein language model" to date.
This model is actually a continuation of the ESM Fold protein prediction model released by Meta in July this year.
When ESMFold was originally released, it was already on par with mainstream protein models such as AlphaFold2 and RoseTTAFold. But the prediction speed of ESMFold is an order of magnitude faster than AlphaFold2!
It may be difficult to understand the speed comparison between the three by talking about the order of magnitude. Just look at the picture below to understand.
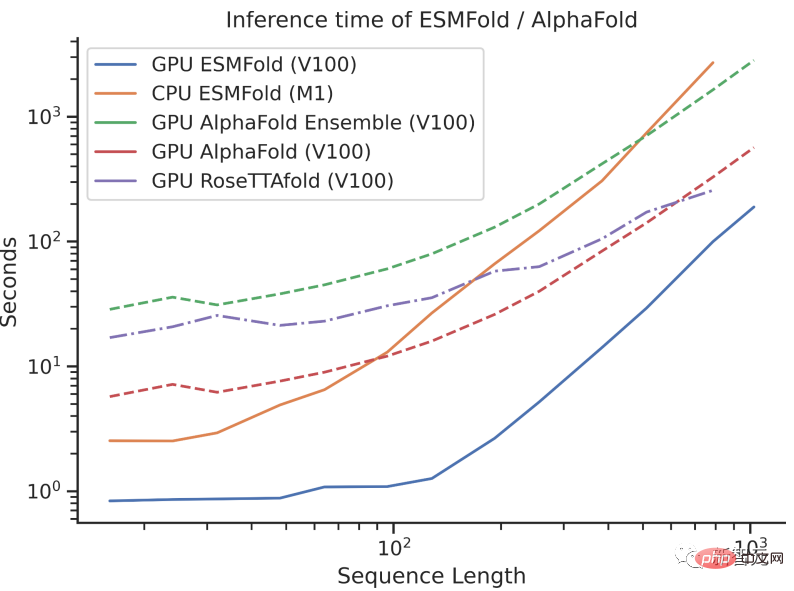
#The release of the ESM Atlas database has given the large language model with 15 billion parameters the widest range of uses.
This allows scientists to search and analyze previously uncharacterized structures on a scale of hundreds of millions of proteins and discover new proteins useful in medicine and other applications.
Language model is really "universal"
Just like text, proteins can also be written as character sequences.
Among them, each "character" that constitutes a protein corresponds to one of the 20 standard chemical elements-amino acid. And each amino acid has different properties.
But it is a big challenge to understand this "biological language".
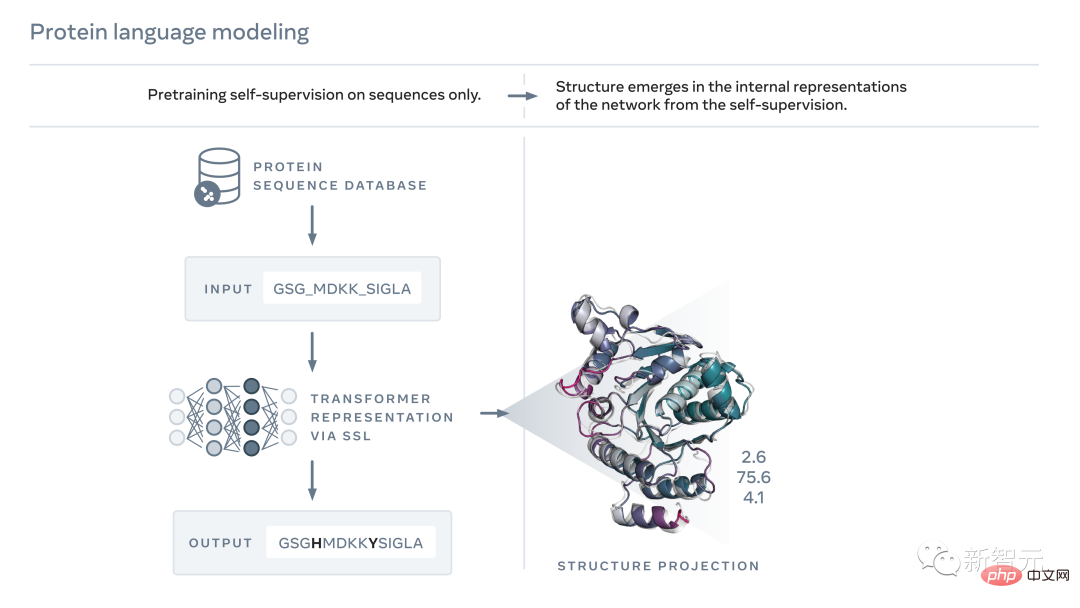
Although, as just said, both a protein sequence and a piece of text can be written as characters, there are profound and fundamental differences between them.
On the one hand, the number of different combinations of these "characters" is an astronomical number. For example, for a protein composed of 200 amino acids, there are 20^200 possible sequences, more than the number of atoms in the currently explorable universe.
On the other hand, each sequence of amino acids folds into a three-dimensional shape according to the laws of physics. Moreover, not all sequences fold into coherent structures; many fold into disordered forms, but it is this elusive shape that determines the protein's function.
For example, if a certain amino acid appears at one position, this amino acid usually pairs with an amino acid at another position. Then, they are likely to interact in the subsequent folded structure.
Artificial intelligence can learn and read these patterns by observing protein sequences, and then infer the actual structure of the protein.
In 2019, Meta presented evidence that language models learn the properties of proteins, such as their structure and function.

Paper address: https://www.pnas.org/doi/10.1073/pnas.2016239118
Using masks as a form of self-supervised learning The trained model can correctly fill in the gaps in a paragraph of text, such as "Do you want __, this is ________".
With this method, Meta trained a language model based on millions of natural protein sequences, thereby filling gaps in protein sequences, such as "GL_KKE_AHY_G".
Experiments show that this model can be trained to discover information about the structure and function of proteins.
In 2020, Meta released ESH1b, the most advanced protein language model at the time, which has been used in various applications, including helping scientists predict the evolution of the new coronavirus and discover the causes of genetic diseases.
Paper address: https://www.biorxiv.org/content/10.1101/2022.08.25.505311v1
Now, Meta has expanded this approach At scale, the next-generation protein language model ESM-2 was created, a large model with 15 billion parameters.
As the model scales from 8 million parameters to 15 million parameters, information emerging from the internal representation enables three-dimensional structure predictions at atomic resolution.
Understand the "protein language" and make life more transparent
Since billions of years ago, the evolution of living things has formed a protein language. Language can form complex and dynamic molecular machines out of simple building blocks. Learning to read the language of proteins is an important step in our understanding of the natural world.
AI can provide us with new tools to understand the natural world. Just like a microscope, it allows us to observe the world at an almost infinitesimal scale and opens up a new understanding of life. AI can help us understand the vast range of diversity in nature and see biology in a new way.
Currently, most AI research is about allowing computers to understand the world in a way similar to humans. The language of proteins is incomprehensible to humans and even to the most powerful computational tools.
So, the significance of Meta’s work is to reveal the huge advantages of AI when crossing fields, namely: large language models that have made progress in machine translation, natural language understanding, speech recognition and image generation, Also able to learn profound information about biology.
This time, Meta makes this work public, shares data and results, and builds on the insights of others. We hope that the release of this large-scale structural atlas and rapid protein folding model can promote further scientific progress and make us better understand the world around them.
References:
https://ai.facebook.com/blog/protein-folding-esmfold-metagenomics/?utm_source=twitter&utm_medium=organic_social&utm_campaign=blog
The above is the detailed content of Meta creates the first 'Protein Universe' panorama! Using a 15 billion parameter language model, more than 600 million protein structures were predicted. For more information, please follow other related articles on the PHP Chinese website!
Related articles
See more- Technology trends to watch in 2023
- How Artificial Intelligence is Bringing New Everyday Work to Data Center Teams
- Can artificial intelligence or automation solve the problem of low energy efficiency in buildings?
- OpenAI co-founder interviewed by Huang Renxun: GPT-4's reasoning capabilities have not yet reached expectations
- Microsoft's Bing surpasses Google in search traffic thanks to OpenAI technology

