 Technology peripherals
Technology peripherals AI
AI Shi Yigong and other teams appear on the cover of Science: AI and cryo-electron microscopy reveal the 'atomic level' NPC structure, a breakthrough in life sciences
Shi Yigong and other teams appear on the cover of Science: AI and cryo-electron microscopy reveal the 'atomic level' NPC structure, a breakthrough in life sciencesBefore starting the text, let’s take a look at a picture. In the picture below, it is obvious that the right half of the picture represents richer information and a clearer structure. The left half of the picture in 2016 has a relatively simple structure and represents less information:

In fact, what is shown above is an image of the nuclear pore complex (NPC). The nuclear pore complex, composed of approximately 1,000 protein subunits, is responsible for the busy transport of macromolecules between the nucleus and cytoplasm of eukaryotic cells. It is also its only two-way channel connecting the cytoplasm and nucleus. In addition to coordinating transport, NPCs organize essential life events such as transcription, mRNA maturation, spliceosome and ribosome assembly. The powerful role of NPC has become a key point in disease mutation and host-pathogen interactions.
Thanks to the development of low-resolution whole nuclear pore structure and high-resolution nuclear pore composition structure technology, cell nuclear pores have received more and more attention. However, using this information to correctly assemble more than 30 different protein copies and construct high-resolution three-dimensional structures has been a difficult challenge.
Today, "Science" magazine published 5 papers as cover features, 3 of which jointly revealed the near-atomic resolution cryo-electron microscopy structure of the human nuclear pore complex. Two other studies presented single-particle cryo-EM images of the vertebrate nuclear pore complex in Xenopus laevis. This cover article stitches together multiple studies to create a nearly atomic-level picture of human NPCs.

Paper address: https://www.science.org/doi/pdf/10.1126/science.add2210
The findings build on decades of research including biochemical reconstructions, X-ray crystallography, mass spectrometry, mutagenesis and cell biology. Human NPCs were reconstructed using greatly improved cryo-electron tomography and components were accurately modeled using artificial intelligence techniques. There are other studies that have improved the resolution of single-particle cryo-EM, enabling the visualization of secondary structural elements and residue-level details of vertebrate NPCs. The molecular assemblage enriches our understanding of the architecture of vertebrate and human NPCs—from the old nuclear scaffolding to the connexins that hold the parts together, and from the nuclear membrane anchoring to the cytoplasmic filaments above the central transport channel.
The research results reported here represent a win-win cooperation between experimental structural biology and artificial intelligence, and are another victory for mankind to explore the biological microscopic world. In addition, it also demonstrates that the ongoing revolution in resolution is irreplaceable in our quest to understand the principles of construction and design of macromolecular assemblies.
Below is a cross-sectional view of the human nuclear pore complex in 2022, with newly resolved components including the symmetric core (orange) and cytoplasmic filaments (yellow):

Five research papers
Paper 1: "Architecture of the cytoplasmic face of the nuclear pore"

Paper address: https://www.science.org/doi/10.1126/science.abm9129
The nuclear pore complex (NPC) is the only bidirectional channel for nucleocytoplasmic transport. Despite recent progress in elucidating the symmetric core structure of the NPC, the asymmetrically distributed cytoplasmic surface that is a hotspot for mRNA export and nucleoporin-related diseases remains elusive.
Researchers from Caltech and other institutions report a composite structure of the human cytoplasmic plane obtained by combining biochemical reconstruction, crystal structure determination, cryo-electron tomography reconstruction and physiological verification. While species-specific motifs anchor an evolutionarily conserved, ~540 kilodalton heterohexameric cytoplasmic filament nucleoporin complex above the central transport channel, attachment of the NUP358 pentameric bundle depends Bicyclic arrangement in the coat nucleoporin complex. The complex structures they reveal and their predictive power provide a rich basis for elucidating the molecular basis of mRNA export and nucleoporin diseases.

Cytoplasmic face of human NPC.
##Paper 2: "Architecture of the linker-scaffold in the nuclear pore"

Paper address: https://www.science.org/doi/10.1126/science.abm9798
Although The arrangement of structured scaffolding nucleoporins in the symmetric core of NPCs has been determined, but their cohesion through multivalent unstructured linker nucleoporins remains elusive.
By combining biochemical reconstruction, high-resolution structure determination, cryo-electron tomography reconstruction, and physiological validation, Caltech researchers elucidated the evolutionarily conserved joint-scaffold structure that resulted in The human NPC has a near-atomic composite structure core with approximately 64 megadalton symmetry. While joints typically serve a rigid role, the NPC's joint scaffold provides the necessary plasticity and robustness for the reversible contraction and expansion of its central transport channels and the emergence of lateral channels. Their results significantly advance the structural characterization of the NPC symmetry core and lay the foundation for future functional studies.

Joint scaffolding structure of the human NPC symmetry core.
Paper 3: "AI-based structure prediction empowers integrative structural analysis of human nuclear pores"

Paper address: https://www.science.org/doi/10.1126/science.abm9506
Although nuclear pore complexes (NPCs) mediate nucleocytoplasmic transport, their intricate 120-megadalton architecture remains incompletely understood. Researchers at the Max Planck Institute for Biophysics and others report a 70-megadalton model of a human NPC scaffold with explicit membranes and multiple conformational states.
They combine AI-based structural predictions with in situ and cellular cryo-electron tomography, comprehensive modeling. The results show that linker nucleoporins organize scaffolds within and between subcomplexes to build higher-order structures. Microsecond-long molecular dynamics simulations show that the scaffold is not required to stabilize the fusion of the inner and outer nuclear membranes, but rather to enlarge the central pore. They illustrate how AI-based modeling can be combined with in situ structural biology to understand subcellular structures across levels of spatial organization.

70 MegaDalton model of the human NPC scaffold architecture.
Paper 4: "Structure of the cytoplasmic ring of the Xenopus laevis nuclear pore complex"

Paper address: https://www.science.org/doi/10.1126/science.abl8280
Westlake University and Tsinghua University Single-particle cryo-electron microscopy reconstruction of the cytoplasmic ring subunit of Xenopus laevis NPC at 3.7-4.7 angstrom resolution. Of these, the structure of the amino-terminal domain of Nup358 was solved to 3.0 Å, which facilitated the identification of five Nup358 molecules in each cytoplasmic ring subunit.
The researchers' final model of the cytoplasmic ring subunit includes five Nup358, two Nup205, and two Nup93 molecules, as well as two previously characterized Y complexes. The carboxyl-terminal fragment of Nup160 serves as an organizing center for the apex of each Y complex. Structural analysis reveals how Nup93, Nup205, and Nup358 promote and enhance the assembly of a cytoplasmic ring scaffold formed primarily by two layers of Y complexes.

## Cryo-EM structure of the bilayered cytoplasmic ring of Xenopus laevis NPC.
Paper 5: "Structure of cytoplasmic ring of nuclear pore complex by integrative cryo-EM and AlphaFold"

Paper address: https://www.science.org/doi/10.1126/science.abm9326
Researchers from Harvard Medical School and other institutions used single-particle cryo-electron microscopy and AlphaFold prediction to determine a nearly complete NPC cytoplasmic ring structure from Xenopus laevis oocytes. Specifically, they used AlphaFold to predict the structure of nucleoporins and fit the medium-resolution map using prominent secondary structure density as a guide.
Additionally, certain molecular interactions were further established or confirmed through complex predictions using AlphaFold. The researchers identified five binding modes for Nup358, the largest NPC subunit with Phe-Gly repeats for transport. They predicted that Nup358 contains a coiled-coil domain that provides activity to help it serve as a nucleation center for NPC formation under certain conditions.

## Cryo-EM structure of Xenopus NPC cytoplasmic loops.
The above is the detailed content of Shi Yigong and other teams appear on the cover of Science: AI and cryo-electron microscopy reveal the 'atomic level' NPC structure, a breakthrough in life sciences. For more information, please follow other related articles on the PHP Chinese website!
 Are You At Risk Of AI Agency Decay? Take The Test To Find OutApr 21, 2025 am 11:31 AM
Are You At Risk Of AI Agency Decay? Take The Test To Find OutApr 21, 2025 am 11:31 AMThis article explores the growing concern of "AI agency decay"—the gradual decline in our ability to think and decide independently. This is especially crucial for business leaders navigating the increasingly automated world while retainin
 How to Build an AI Agent from Scratch? - Analytics VidhyaApr 21, 2025 am 11:30 AM
How to Build an AI Agent from Scratch? - Analytics VidhyaApr 21, 2025 am 11:30 AMEver wondered how AI agents like Siri and Alexa work? These intelligent systems are becoming more important in our daily lives. This article introduces the ReAct pattern, a method that enhances AI agents by combining reasoning an
 Revisiting The Humanities In The Age Of AIApr 21, 2025 am 11:28 AM
Revisiting The Humanities In The Age Of AIApr 21, 2025 am 11:28 AM"I think AI tools are changing the learning opportunities for college students. We believe in developing students in core courses, but more and more people also want to get a perspective of computational and statistical thinking," said University of Chicago President Paul Alivisatos in an interview with Deloitte Nitin Mittal at the Davos Forum in January. He believes that people will have to become creators and co-creators of AI, which means that learning and other aspects need to adapt to some major changes. Digital intelligence and critical thinking Professor Alexa Joubin of George Washington University described artificial intelligence as a “heuristic tool” in the humanities and explores how it changes
 Understanding LangChain Agent FrameworkApr 21, 2025 am 11:25 AM
Understanding LangChain Agent FrameworkApr 21, 2025 am 11:25 AMLangChain is a powerful toolkit for building sophisticated AI applications. Its agent architecture is particularly noteworthy, allowing developers to create intelligent systems capable of independent reasoning, decision-making, and action. This expl
 What are the Radial Basis Functions Neural Networks?Apr 21, 2025 am 11:13 AM
What are the Radial Basis Functions Neural Networks?Apr 21, 2025 am 11:13 AMRadial Basis Function Neural Networks (RBFNNs): A Comprehensive Guide Radial Basis Function Neural Networks (RBFNNs) are a powerful type of neural network architecture that leverages radial basis functions for activation. Their unique structure make
 The Meshing Of Minds And Machines Has ArrivedApr 21, 2025 am 11:11 AM
The Meshing Of Minds And Machines Has ArrivedApr 21, 2025 am 11:11 AMBrain-computer interfaces (BCIs) directly link the brain to external devices, translating brain impulses into actions without physical movement. This technology utilizes implanted sensors to capture brain signals, converting them into digital comman
 Insights on spaCy, Prodigy and Generative AI from Ines MontaniApr 21, 2025 am 11:01 AM
Insights on spaCy, Prodigy and Generative AI from Ines MontaniApr 21, 2025 am 11:01 AMThis "Leading with Data" episode features Ines Montani, co-founder and CEO of Explosion AI, and co-developer of spaCy and Prodigy. Ines offers expert insights into the evolution of these tools, Explosion's unique business model, and the tr
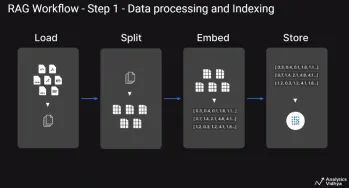 A Guide to Building Agentic RAG Systems with LangGraphApr 21, 2025 am 11:00 AM
A Guide to Building Agentic RAG Systems with LangGraphApr 21, 2025 am 11:00 AMThis article explores Retrieval Augmented Generation (RAG) systems and how AI agents can enhance their capabilities. Traditional RAG systems, while useful for leveraging custom enterprise data, suffer from limitations such as a lack of real-time dat


Hot AI Tools

Undresser.AI Undress
AI-powered app for creating realistic nude photos

AI Clothes Remover
Online AI tool for removing clothes from photos.

Undress AI Tool
Undress images for free

Clothoff.io
AI clothes remover

Video Face Swap
Swap faces in any video effortlessly with our completely free AI face swap tool!

Hot Article

Hot Tools

MantisBT
Mantis is an easy-to-deploy web-based defect tracking tool designed to aid in product defect tracking. It requires PHP, MySQL and a web server. Check out our demo and hosting services.

Dreamweaver Mac version
Visual web development tools

SublimeText3 Mac version
God-level code editing software (SublimeText3)

PhpStorm Mac version
The latest (2018.2.1) professional PHP integrated development tool

WebStorm Mac version
Useful JavaScript development tools





