Home >Technology peripherals >AI >To predict protein-DNA binding specificity, USC team develops new geometric deep learning method
To predict protein-DNA binding specificity, USC team develops new geometric deep learning method
- PHPzOriginal
- 2024-08-19 13:34:101315browse
Editor | Radish Skin
Predicting protein-DNA binding specificity is a challenging but crucial task that helps understand gene regulation. Protein-DNA complexes typically bind to selected DNA targets, whereas proteins bind to a broad range of DNA sequences with varying degrees of binding specificity. This information is not directly accessible in a single structure.
To obtain this information, researchers from the University of Southern California and the University of Washington proposed the Deep Binding Specificity Predictor (DeepPBS), a geometric deep learning model designed to Prediction of binding specificity based on protein-DNA structure.
DeepPBS can extract interpretable protein heavy atom importance scores of interface residues. These scores were validated by mutagenesis experiments when aggregated at the protein residue level. Applied to designed proteins targeting specific DNA sequences, DeepPBS was shown to predict experimentally measured binding specificities.
The study was titled "Geometric deep learning of protein–DNA binding specificity" and was published in "Nature Methods" on August 5, 2024.

Transcription factors regulate life processes by binding to specific DNA sequences. This binding mechanism includes electrostatic interactions, deoxyribose stacking effects, and the formation of hydrogen bonds.
Protein-DNA structural information is usually obtained through experimental methods such as X-ray crystallography, nuclear magnetic resonance spectroscopy or cryo-electron microscopy, and is stored in the Protein Data Bank (PDB). These structures typically demonstrate bound DNA sequences and their physicochemical interactions, but do not cover all possible binding sequences.
On the other hand, high-throughput experiments such as protein binding microarrays, SELEX-seq, etc. are able to capture the range of potential binding sequences but lack structural information.
Therefore, combining structural data and high-throughput experimental data is crucial to fully understand the binding specificity of transcription factors.
Currently, predicting the binding specificity of a specific protein sequence within a protein family remains a challenging and unsolved problem. This difficulty is exacerbated by structural changes in the context and the vast mechanistic diversity.
"The structure of a protein-DNA complex contains proteins that typically bind to a single DNA sequence. In order to understand gene regulation, it is important to understand the binding specificity of a protein to any DNA sequence or genomic region." said Professor Remo Rohs of the University of Southern California. .
In the latest study, researchers from the University of Southern California and the University of Washington introduced Binding Specificity Deep Predictors (DeepPBS).
Rohs explained: "DeepPBS is an artificial intelligence tool that replaces high-throughput sequencing or structural biology experiments to reveal protein-DNA binding specificity."
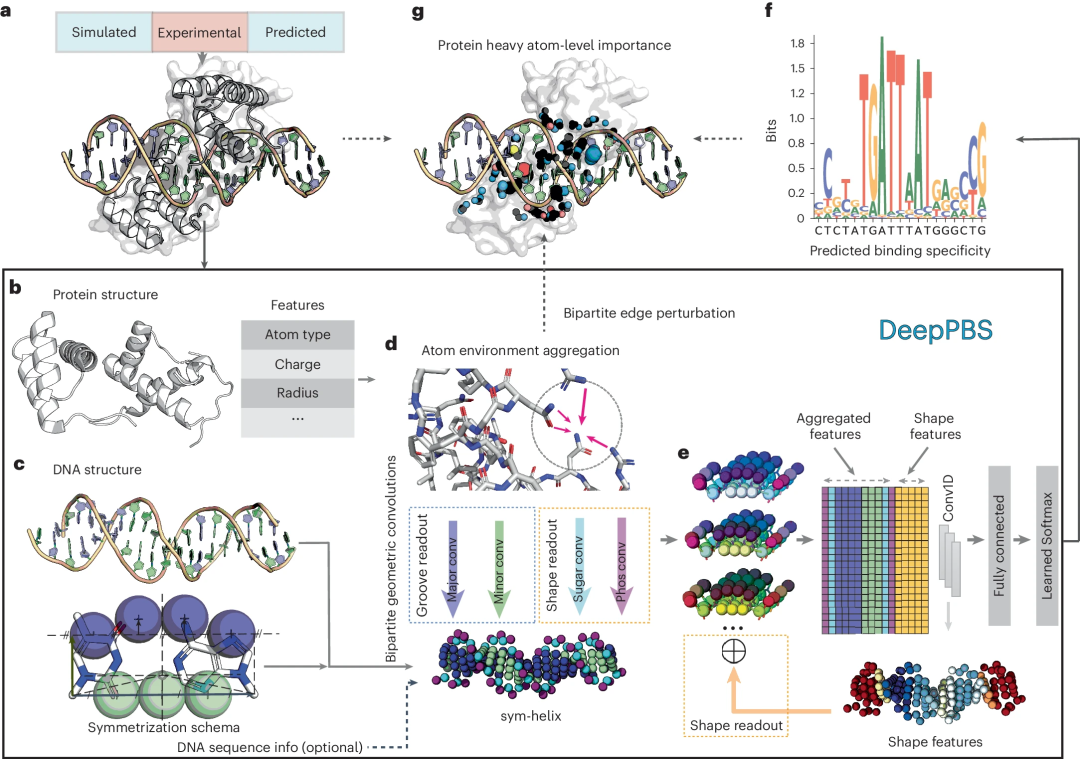
This deep learning model aims to capture the physicochemical and geometric context of protein-DNA interactions to predict binding specificity, expressed as a position weight matrix (PWM) based on a given protein-DNA structure. DeepPBS functions across protein families and serves as a bridge between structure determination and binding specificity determination experiments.
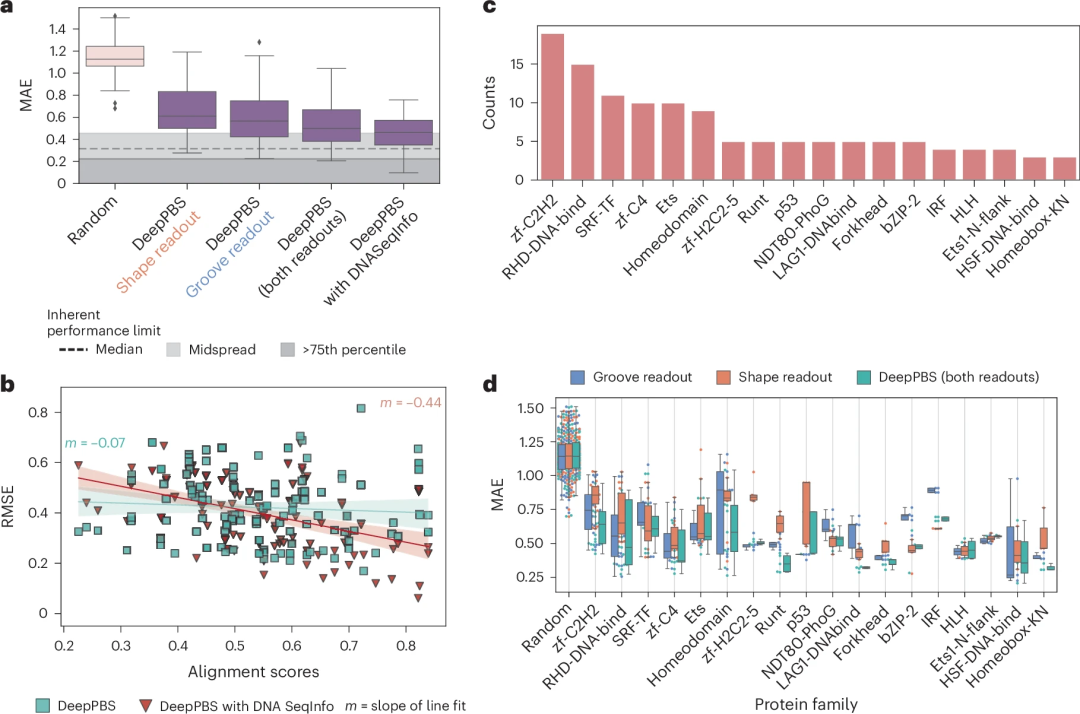
Illustration: Performance of DeepPBS for predicting binding specificity across protein families. (Source: paper)
Inputs to DeepPBS are not limited to experimental structures. The rapid development of protein structure prediction methods, including AlphaFold, OpenFold, and RoseTTAFold, and protein-DNA complex modelers, such as RoseTTAFoldNA (RFNA), RoseTTAFold All-Atom, MELD-DNA, and AlphaFold3, has resulted in structural data available for analysis The numbers are growing exponentially.
This scenario highlights the growing need for universal computational models for analyzing protein-DNA structures. The researchers demonstrate how DeepPBS can be used in conjunction with structure prediction methods to predict the specificity of proteins for which no experimental structure is available.
Additionally, the design of protein-DNA complexes can be improved by using DeepPBS feedback to optimize binding to DNA. The researchers show that this pipeline performs comparably to a recent family-specific model, rCLAMPS, while being more general: Specifically, DeepPBS is not restricted by protein families, can handle biological assemblies, and can predict DNA side chain preferences.
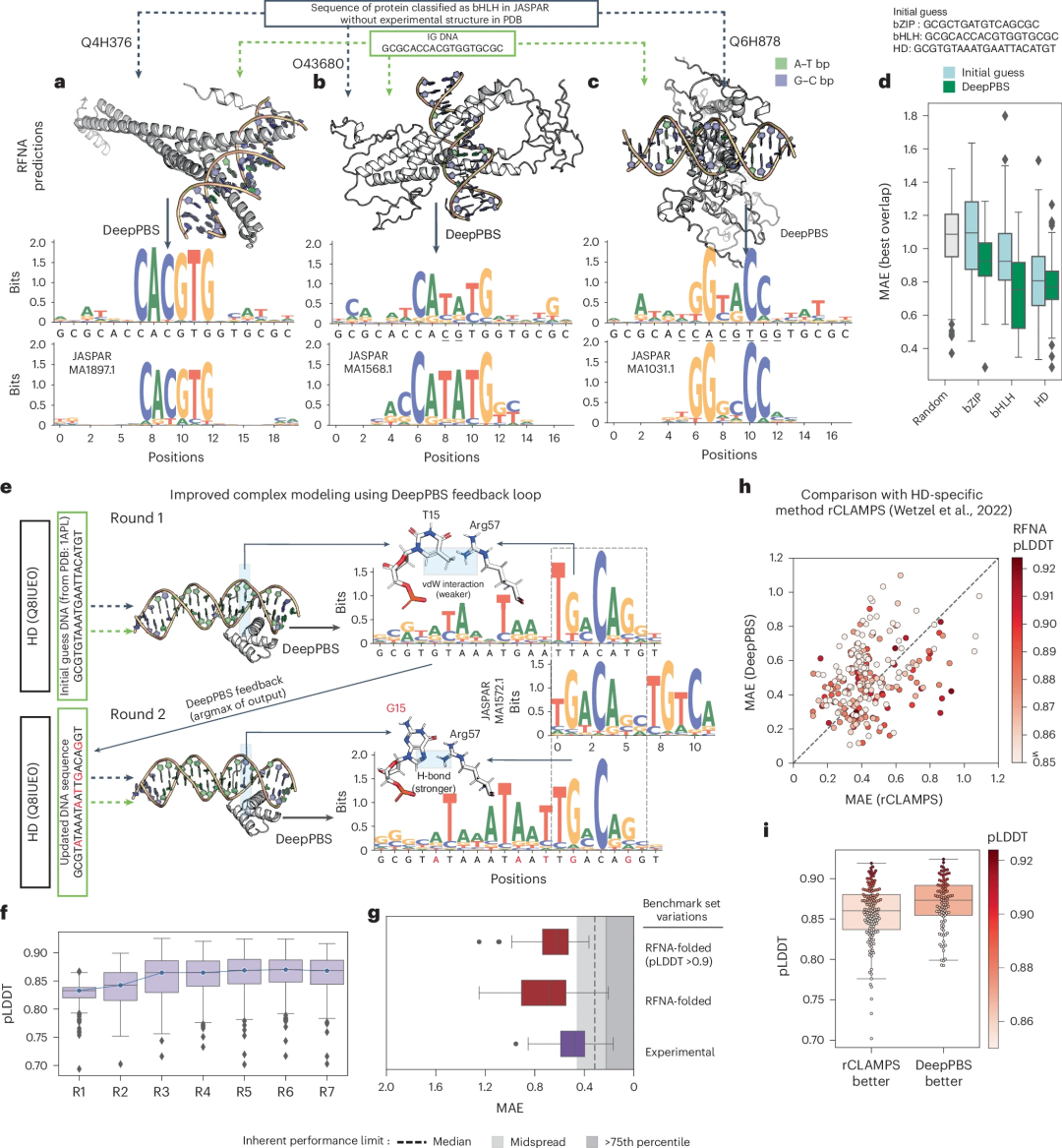
Illustration: Application of DeepPBS in predicting the structure of protein-DNA complexes. (Source: paper)
In terms of interpretability, the “relative importance” (RI) scores of different heavy atoms in proteins interacting with DNA can be extracted from DeepPBS.
As a case study of proteins important for cancer development, the researchers analyzed the p53-DNA interface through these RI scores and linked them to existing literature for validation.
Also, DeepPBS scores agree well with existing knowledge and can be aggregated to produce reasonable agreement with alanine scanning mutagenesis experiments.
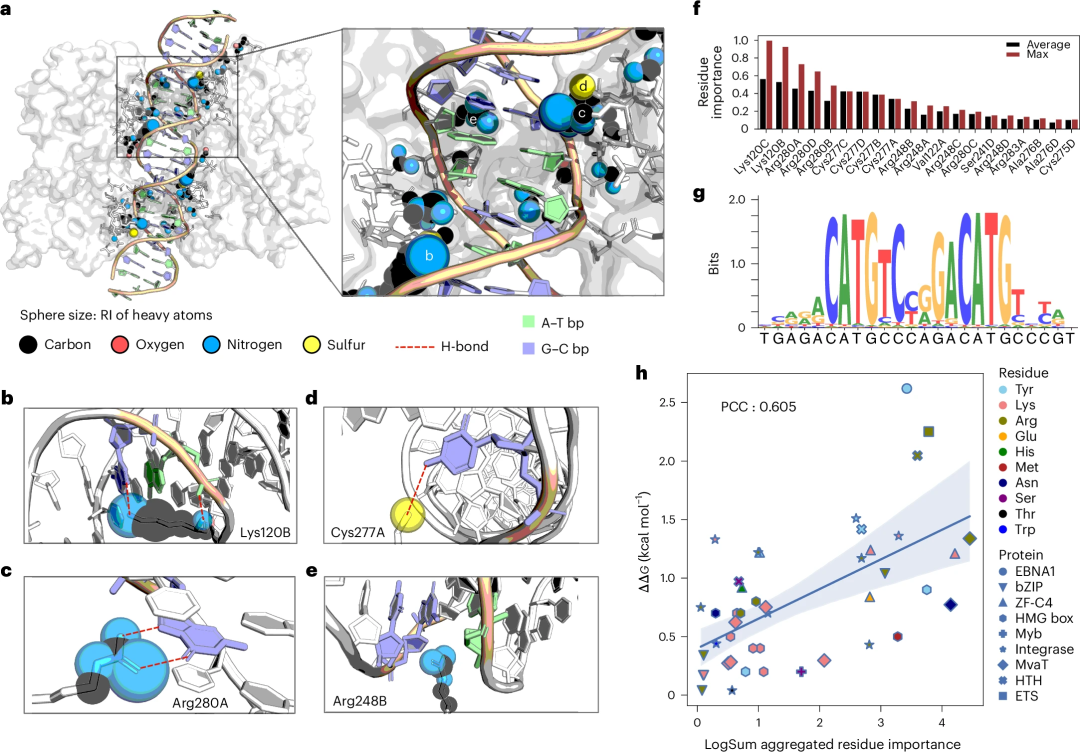
Illustration: Taking the visualization of DeepPBS importance scores in the p53-DNA interface as an example to study and conduct experimental verification. (Source: paper)
In additional proof-of-principle studies, researchers applied DeepPBS to in silico-designed protein-DNA complexes targeting specific DNA sequences from a recent experiment combining structural design and DNA mutagenesis research. DeepPBS can also be used to analyze molecular simulation trajectories.
"It is important for researchers to find a method that works for all proteins and is not limited to a well-studied protein family. This method also allows us to design new proteins." Rohs said.
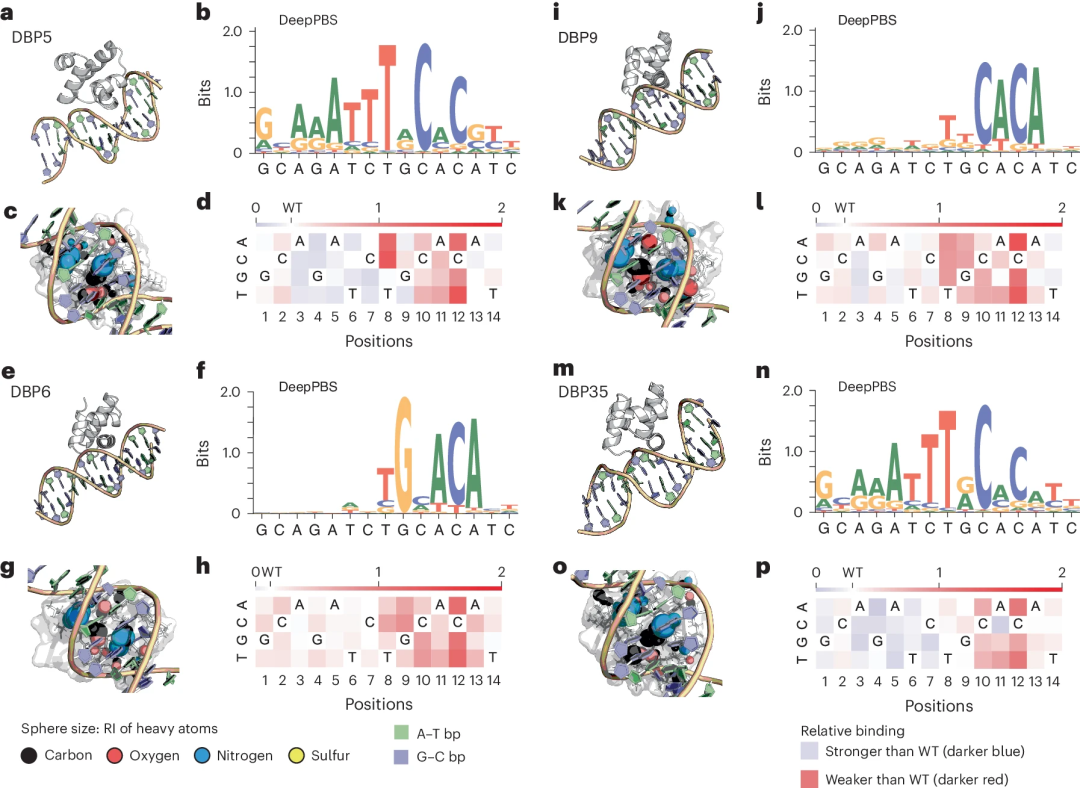
Illustration: Applying DeepPBS to in silico designed HTH scaffolds targeting specific DNA sequences. (Source: paper)
The current version of DeepPBS has inherent limitations. It is tailored for double-stranded DNA and does not yet work with single-stranded DNA, RNA, or chemically modified bases.
However, the model has the potential to be extended to accommodate these different scenarios as well as other polymer-polymer interactions, and potentially to mechanistic mutations. The DeepPBS architecture can be optimized and extended in terms of application and engineering enhancements.
Nevertheless, Rohs said DeepPBS will have a wide range of applications. This new research approach may accelerate the design of new drugs and treatments that target specific mutations in cancer cells, as well as lead to new discoveries in synthetic biology and applications in RNA research.
DeepPBS: https://deeppbs.usc.edu
The above is the detailed content of To predict protein-DNA binding specificity, USC team develops new geometric deep learning method. For more information, please follow other related articles on the PHP Chinese website!
Related articles
See more- Technology trends to watch in 2023
- How Artificial Intelligence is Bringing New Everyday Work to Data Center Teams
- Can artificial intelligence or automation solve the problem of low energy efficiency in buildings?
- OpenAI co-founder interviewed by Huang Renxun: GPT-4's reasoning capabilities have not yet reached expectations
- Microsoft's Bing surpasses Google in search traffic thanks to OpenAI technology

