 Technology peripherals
Technology peripherals AI
AI With 130,000 annotated neurons and 53 million synapses, Princeton University and others released the first complete 'adult fly' brain connectome
With 130,000 annotated neurons and 53 million synapses, Princeton University and others released the first complete 'adult fly' brain connectomeWith 130,000 annotated neurons and 53 million synapses, Princeton University and others released the first complete 'adult fly' brain connectome
As of today, there are many projects that have mapped the whole-brain connectome of various organisms, from Caenorhabditis elegans (302 neurons) to Drosophila melanogaster (~100,000 neurons). Drosophila melanogaster is one of the most thoroughly studied organisms by humans. As of 2017, eight Nobel Prizes have been awarded to research using fruit flies.
Researchers’ research on Drosophila continues. Recently, researchers from Princeton University and other institutions released the Drosophila whole-brain connectome, including about 130k annotated neurons. and tens of millions of types of synapses.

Everyone knows to some extent that a basic nervous system has existed since ancient animals, but the emergence of the brain system can be traced back to 500 million years ago . Research shows that dividing the brain into different regions helps understand its function.
However, for many years, there has been controversy about neural connection maps at the neuronal and synaptic level. The main reason for this phenomenon is that humans lack the ability to reconstruct such connection maps. technology. Things only started to change in the early 21st century as technology evolved.
Until today, researchers from Princeton University and other institutions have released the Drosophila whole-brain connectome, which is the first complete neural connection map of the adult Drosophila brain.
 Picture
Picture
Paper address: https://www.biorxiv.org/content/10.1101/2023.06.27.546656v1. full.pdf
 ##Picture
##Picture
After the release of the results, someone said: "Many projects have mapped the full range of various organisms. Map of the brain connectome, from Caenorhabditis elegans (302 neurons) to Drosophila melanogaster (~100k neurons). With our current computing power, why can't we perform accurate computer simulations of these organisms in a virtual 3D environment? 》
The first complete neural connection map of the adult fruit fly brain
The brain of fruit flies looks very small, with 10^5 Neurons and 10^8 synapses, nevertheless, fruit flies use these to see, smell, hear, walk, and of course fly.
For a long time, researchers have reconstructed parts of the Drosophila brain through electron microscope (EM) images. These images have good clarity and can show the small size of neurons. Branches and connected synapses. The resulting connectivity map of neural circuits provides key insights into how the brain generates social behaviors, memory-related behaviors, and navigation behaviors.
However, although the EM method has been applied to some areas of the brain and reconstructed information-rich local connection maps, this method is still insufficient for a more comprehensive understanding of brain function. insufficient.
Previously, researchers built a single synapse grid based on the research of Ito et al. The grid used in this article is based on the JFRC2 standard brain template that previously generated complete brain segmentation. These Grids are also used in the Virtual Fly Brain project. This study moves these meshes from JFRC2 space into FlyWire (FAFB14.1) space through a series of non-rigid transformations.
Note: FlyWire is a whole-brain connectomics platform for exploring the Drosophila brain. Since 2019, scientists and experienced proofreaders have been using FlyWire to proofread AI segmentations of whole fruit fly brains. As of June 2023, more than 120,000 neurons have been proofread in FlyWire, including the entire central brain.
As shown below, the entire adult Drosophila brain reconstructed in this study contains 127,978 neurons (Figure 1a) with 53 million synapses between them. Whole-brain images of adult female Drosophila melanogaster (Fig. 1e, f) were previously obtained by serial section transfer EM and released into the public domain by Zheng et al.
 picture
picture
This article shows that the connection map obtained by reconstructing the entire Drosophila brain in this study is complete enough to be called a "connectome." The connectome is significantly improved compared to Caenorhabditis elegans (300 neurons, less than 10^4 synapses) and Drosophila first instar larvae (3,000 neurons, 5×10^5 synapses) A leap forward: the connectome not only exceeds half of the Drosophila brain in quantity, it also covers the subesophageal zone (SEZ) of the Drosophila central brain, which is very important for taste and mechanical perception. In addition, the connectome also Covers the processes that drive motor neurons from the Drosophila brain down.
The figure below shows the categories of neurons. Figure (a) shows the classification of neurons in the Drosophila brain by "stream": intrinsic, afferent, and efferent. Each flow class is then further divided into "superclasses" based on location and function. The first publicly released compound eye was missing about 8,000 retinal cells and four tiny eyeballs in one hemisphere, parts indicated by hatched bars. (b) Using these neuronal annotations, the study created an aggregated synaptic map between superclasses in the Drosophila brain. (c) Rendering of all neurons in each superclass. (d) In addition to the ophthalmic nerve and cervical connective tissue (CV), there are 8 nerves in each hemisphere. All neurons that cross the nerve are reconstructed and explained. (e) Sensory neurons can be subdivided based on their responses to sensory modalities. In FlyWire, almost all sensory neurons have been classified by modality. (f) Rendering of all non-visual sensory neurons.
 ##Picture
##Picture
The following picture is a diagram of the fruit fly brain responsible for processing visual information:
 Picture
Picture
The picture below shows the flow of information through the central brain of a fruit fly:
 Picture
Picture
The largest ever validated collection of cell types
The FlyWire connectome is the largest and most complex connectome ever obtained. In another paper, "A consensus cell type atlas from multiple connectomes reveals principles of circuit stereotypy and variation," the team raised and answered some key questions to further explain connectomes of this scale:
1) How to know which edges are important?
2) How to simplify connectome diagrams to aid automated or manual analysis?
3) To what extent is this connectome a snapshot of a single brain or representative of the species as a whole?
 Picture
Picture
Paper link: https://www.biorxiv.org/content/10.1101/2023.06.27.546055v1. full.pdf
These questions are inextricably linked to connectome annotation and cell type identification within and across data sets.
At the most basic level, navigating this connectome would be very challenging without a comprehensive annotation system. So, the team's annotations for this paper provide an indexed and hierarchical list of human-readable parts, allowing biologists to explore the systems and neurons of interest to them.
Connectome annotation is also crucial to ensure data quality, as it inevitably reveals segmentation errors that must be corrected. Furthermore, Drosophila has a rich history of exploring the circuit basis of a wide range of innate and acquired behaviors, as well as their developmental genetic origins; realizing the full potential of the dataset will only be possible by cross-identifying cell types within the connectome with those previously identified in Characterization in published and ongoing literature.
Comparison with partial hemibrain connectomes confirms that most fly cell types are highly stereotyped; and that connections within simply defined and general heuristic connectomes are reliable and more likely to be functional sexual. However, this also revealed unexpected changes in some cell types and showed that many cell types originally reported in hemibrains cannot be reliably reidentified. This finding led to the need to develop and apply a new and powerful method to define cell types across connectomics data sets.
In this study, the researchers generated human-readable data at different levels of granularity (superclass, cell class, lineage class, etc.) for all neurons in the fly brain. Note.
The map of 4,179 cell types provided by the researchers is not the largest ever (5,620 in one half of the brain, and recent work in mouse brains provided as many as 5,000 molecular clusters) . However, it is by some means the largest validated collection of cell types ever assembled.
Cell type is a provable hypothesis about biological variability within and across animals. In C. elegans , 118 cell types inferred from the initial connectome have been unequivocally supported by subsequent analysis of connectome and molecular data. In some mammals, generating catalogs of 100 cell types is possible and has been validated with multimodal data such as in the retina or motor cortex.
However, large-scale molecular maps yield highly informative hierarchies, but no attempt has been made to precisely define terminal cell types—the finest units across individuals. The researchers tested this falsifiable cell type hypothesis for the first time on more than 5,000 cell types, confirming or correcting approximately 3,166 cell types in connectome data from 3 hemispheres.
It is worth noting that connectome data is particularly meaningful for cell type delineation: it is itself multimodal (by providing morphology and connectivity), and it can see All cells within the brain (integrity). Cell types that pass this rigorous test are very unlikely to be modified (permanently). Based on this understanding, an additional 850 cell types defined within just two hemispheres of the FlyWire dataset should also be accurate and permanent. The researchers believe that connectome data will become the gold standard for cell types. Therefore, linking molecular and connectomic cell types will be key.
People may be a little surprised or even disappointed that more than a third of half of the brain's cell types cannot yet be identified in FlyWire. The researchers reiterate that most cells can be identified, and they expect to continue to make incremental improvements on current platforms and tools through their own efforts and those of others.
Nevertheless, the current results reveal several important issues: first, many of the cell types identified using data from a single brain hemisphere now need to be modified; second, new multi-connectivity The component typing method (Figure 6) provides a powerful and efficient approach to this problem; third, examples of continuous variation in adult flies are often associated with mammalian cell types, and researchers now also have the tools and data to perform methods that were previously impossible. accuracy to handle this variation.
 Cross-brain cell typing
Cross-brain cell typing
Overall, this work has laid some foundations for in-depth research on current and the expected normal fly connectome can also facilitate future research on sexual dimorphism, experience-dependent plasticity, whole-brain-scale development, and disease.
The above is the detailed content of With 130,000 annotated neurons and 53 million synapses, Princeton University and others released the first complete 'adult fly' brain connectome. For more information, please follow other related articles on the PHP Chinese website!
 Tesla's Robovan Was The Hidden Gem In 2024's Robotaxi TeaserApr 22, 2025 am 11:48 AM
Tesla's Robovan Was The Hidden Gem In 2024's Robotaxi TeaserApr 22, 2025 am 11:48 AMSince 2008, I've championed the shared-ride van—initially dubbed the "robotjitney," later the "vansit"—as the future of urban transportation. I foresee these vehicles as the 21st century's next-generation transit solution, surpas
 Sam's Club Bets On AI To Eliminate Receipt Checks And Enhance RetailApr 22, 2025 am 11:29 AM
Sam's Club Bets On AI To Eliminate Receipt Checks And Enhance RetailApr 22, 2025 am 11:29 AMRevolutionizing the Checkout Experience Sam's Club's innovative "Just Go" system builds on its existing AI-powered "Scan & Go" technology, allowing members to scan purchases via the Sam's Club app during their shopping trip.
 Nvidia's AI Omniverse Expands At GTC 2025Apr 22, 2025 am 11:28 AM
Nvidia's AI Omniverse Expands At GTC 2025Apr 22, 2025 am 11:28 AMNvidia's Enhanced Predictability and New Product Lineup at GTC 2025 Nvidia, a key player in AI infrastructure, is focusing on increased predictability for its clients. This involves consistent product delivery, meeting performance expectations, and
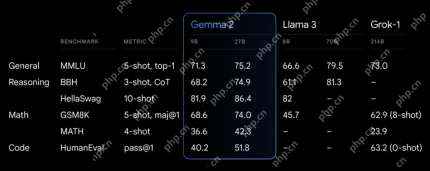 Exploring the Capabilities of Google's Gemma 2 ModelsApr 22, 2025 am 11:26 AM
Exploring the Capabilities of Google's Gemma 2 ModelsApr 22, 2025 am 11:26 AMGoogle's Gemma 2: A Powerful, Efficient Language Model Google's Gemma family of language models, celebrated for efficiency and performance, has expanded with the arrival of Gemma 2. This latest release comprises two models: a 27-billion parameter ver
 The Next Wave of GenAI: Perspectives with Dr. Kirk Borne - Analytics VidhyaApr 22, 2025 am 11:21 AM
The Next Wave of GenAI: Perspectives with Dr. Kirk Borne - Analytics VidhyaApr 22, 2025 am 11:21 AMThis Leading with Data episode features Dr. Kirk Borne, a leading data scientist, astrophysicist, and TEDx speaker. A renowned expert in big data, AI, and machine learning, Dr. Borne offers invaluable insights into the current state and future traje
 AI For Runners And Athletes: We're Making Excellent ProgressApr 22, 2025 am 11:12 AM
AI For Runners And Athletes: We're Making Excellent ProgressApr 22, 2025 am 11:12 AMThere were some very insightful perspectives in this speech—background information about engineering that showed us why artificial intelligence is so good at supporting people’s physical exercise. I will outline a core idea from each contributor’s perspective to demonstrate three design aspects that are an important part of our exploration of the application of artificial intelligence in sports. Edge devices and raw personal data This idea about artificial intelligence actually contains two components—one related to where we place large language models and the other is related to the differences between our human language and the language that our vital signs “express” when measured in real time. Alexander Amini knows a lot about running and tennis, but he still
 Jamie Engstrom On Technology, Talent And Transformation At CaterpillarApr 22, 2025 am 11:10 AM
Jamie Engstrom On Technology, Talent And Transformation At CaterpillarApr 22, 2025 am 11:10 AMCaterpillar's Chief Information Officer and Senior Vice President of IT, Jamie Engstrom, leads a global team of over 2,200 IT professionals across 28 countries. With 26 years at Caterpillar, including four and a half years in her current role, Engst
 New Google Photos Update Makes Any Photo Pop With Ultra HDR QualityApr 22, 2025 am 11:09 AM
New Google Photos Update Makes Any Photo Pop With Ultra HDR QualityApr 22, 2025 am 11:09 AMGoogle Photos' New Ultra HDR Tool: A Quick Guide Enhance your photos with Google Photos' new Ultra HDR tool, transforming standard images into vibrant, high-dynamic-range masterpieces. Ideal for social media, this tool boosts the impact of any photo,


Hot AI Tools

Undresser.AI Undress
AI-powered app for creating realistic nude photos

AI Clothes Remover
Online AI tool for removing clothes from photos.

Undress AI Tool
Undress images for free

Clothoff.io
AI clothes remover

Video Face Swap
Swap faces in any video effortlessly with our completely free AI face swap tool!

Hot Article

Hot Tools

ZendStudio 13.5.1 Mac
Powerful PHP integrated development environment

EditPlus Chinese cracked version
Small size, syntax highlighting, does not support code prompt function

MinGW - Minimalist GNU for Windows
This project is in the process of being migrated to osdn.net/projects/mingw, you can continue to follow us there. MinGW: A native Windows port of the GNU Compiler Collection (GCC), freely distributable import libraries and header files for building native Windows applications; includes extensions to the MSVC runtime to support C99 functionality. All MinGW software can run on 64-bit Windows platforms.

SublimeText3 Chinese version
Chinese version, very easy to use

Notepad++7.3.1
Easy-to-use and free code editor




