 Technology peripherals
Technology peripherals AI
AI Achieving high versatility with small amounts of data, KAIST develops new framework for 3D molecule generation for drug design
Achieving high versatility with small amounts of data, KAIST develops new framework for 3D molecule generation for drug designAchieving high versatility with small amounts of data, KAIST develops new framework for 3D molecule generation for drug design
Editor | Radish Skin
Deep generative models have great potential to accelerate drug design. However, existing generative models often face generalization challenges due to limited data, resulting in less innovative designs.
To address these issues, researchers at KAIST in South Korea proposed an interaction-aware 3D molecular generation functional framework that enables interaction-guided interaction design within the target binding pocket. By utilizing common patterns of protein-ligand interactions as prior knowledge, the model can achieve a high degree of generality with limited experimental data. At the same time, using protein mass-ligand mass as a general pattern for interaction purposes, the model can achieve a good balance between generality and high specificity, which provides generality and predictability for drug design.
The performance of the generated unseen target ligands was comprehensively evaluated by analyzing their binding posture certainty, affinity, diversity and novelty. Furthermore, the efficient design of potential mutation-selective inhibitors demonstrates the applicability of this approach to structure-based drug design.
The study was titled "3D molecular generative framework for interaction-guided drug design" and was published in "Nature Communications" on March 27, 2024.

In data capture and scientific problems, appropriate hierarchical prior knowledge of deep learning models is crucial to developing generalizable models. For example, AlphaFold successfully predicts protein structures by leveraging co-evolutionary information and residue pair representations. Deep generative models are changing the drug design paradigm, but their performance is limited by the lack of activity data on drug molecules, resulting in low generalization capabilities. To improve the performance of deep generative models, we need appropriate prior knowledge to ensure their suitability for generalization of drug molecule activity data, which is critical for predicting challenging compound structures and properties.
Recent generative functional models improve the waveformation capabilities of the model by utilizing the three-dimensional structure of the binding site for structure-based ligand design without relying on activity data. A well-waved model should understand the universal properties of protein-ligand interactions, including hydrogen bonds, salt bridges, hydrophobic interactions, and π-π stacking. This is essential to form a stable binding structure and maintain high affinity. These ubiquitous interaction patterns are the basis for the design of powerful drugs.
Based on these circumstances, KAIST researchers proposed an interaction-aware 3D molecular generation framework. This framework exploits the universal nature of protein-ligand interactions to guide structure-based drug design. The framework consists of two main stages: (1) interaction sensing condition setting and (2) interacting 3D molecule generation.
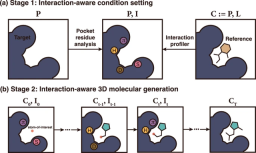
Illustration: Framework concept illustration. (Source: paper)
The first stage of the framework aims to set the interaction conditions I by studying the protein atoms for a given binding site P. The researchers used four types of protein-ligand interactions—hydrogen bonds, salt bridges, hydrophobic interactions, and π-π stacking. Here the researchers only considered the four most dominant interaction types in the Protein Data Bank (PDB), mainly because they used the PDBbind 2020 data set derived from the PDB for model training.
At the same time, the team developed a protein-atom interaction sensing regulation strategy. The researchers define interaction conditions as a one-hot vector of additional interaction types for a set of protein atoms, which indicates whether an atom can participate in a specific interaction and its role in the interaction.
Protein atoms are classified into one of seven categories: anions, cations, hydrogen bond donors and acceptors, aromatic, hydrophobic and non-interacting atoms. Instead of representing the entire interaction information as a single interaction fingerprint, the team's strategy aims to establish interaction conditions locally.
In this work, the researchers mainly determined the interaction categories of bag atoms through two strategies.
During the generation phase, since information on receptor-ligand interactions is not always available, criteria for interaction categories are predefined in order to specify interaction conditions by analyzing each protein atom. This The condition set is called the reference-free interaction condition.
During the training phase, the ground-truth structures of protein-ligand complexes are used to extract interaction conditions.
The researchers also proposed a deep generative model called DeepICL for reverse engineering ligands, which gradually generates atoms in the ligand based on the three-dimensional environment of the pocket and the first-stage interaction conditions.
Although target pockets can form different combinations of protein-ligand interaction types depending on the bound ligand and its binding posture; the team's goal was to reverse engineer one using a 3D conditional generative model called DeepICL. For ligands that satisfy specific interaction combinations, the model can be applied to any type of protein. Researchers use local interaction conditions in the subpockets to which ligands should bind, rather than using the entire interaction information, to prevent undesirable biases toward specific pockets or ligand structures.
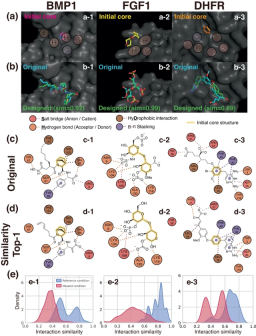
Illustration: Example of interaction-aware conditional ligand elaboration. (Source: Paper)
To demonstrate the framework's ability to perform general structure-based drug design, rather than using typical benchmarks consisting of 105 to 107 computer-generated protein-ligand binding structures, the researchers used only Approximately 104 real crystal structures were selected from the PDBbind database because a good generalization model can successfully extract appropriate features even for small-scale data.
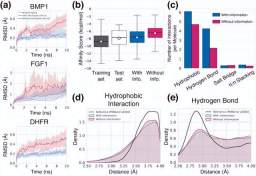
Illustration: Generating the universality of the framework. (Source: Paper)
The researchers evaluated their model by analyzing various aspects of the properties of the generated unseen target ligands—binding stability, affinity, geometric patterning, diversity, and novelty.
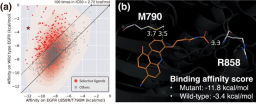
#aIllustration: Modulating selectivity through site-specific interactions controls ligand design. (Source: Paper)
The researchers used the model to solve practical problems where specific interaction sites play a critical role, demonstrating the applicability of their approach to structure-based drug design.
Paper link:https://www.nature.com/articles/s41467-024-47011-2
The above is the detailed content of Achieving high versatility with small amounts of data, KAIST develops new framework for 3D molecule generation for drug design. For more information, please follow other related articles on the PHP Chinese website!
 The AI Skills Gap Is Slowing Down Supply ChainsApr 26, 2025 am 11:13 AM
The AI Skills Gap Is Slowing Down Supply ChainsApr 26, 2025 am 11:13 AMThe term "AI-ready workforce" is frequently used, but what does it truly mean in the supply chain industry? According to Abe Eshkenazi, CEO of the Association for Supply Chain Management (ASCM), it signifies professionals capable of critic
 How One Company Is Quietly Working To Transform AI ForeverApr 26, 2025 am 11:12 AM
How One Company Is Quietly Working To Transform AI ForeverApr 26, 2025 am 11:12 AMThe decentralized AI revolution is quietly gaining momentum. This Friday in Austin, Texas, the Bittensor Endgame Summit marks a pivotal moment, transitioning decentralized AI (DeAI) from theory to practical application. Unlike the glitzy commercial
 Nvidia Releases NeMo Microservices To Streamline AI Agent DevelopmentApr 26, 2025 am 11:11 AM
Nvidia Releases NeMo Microservices To Streamline AI Agent DevelopmentApr 26, 2025 am 11:11 AMEnterprise AI faces data integration challenges The application of enterprise AI faces a major challenge: building systems that can maintain accuracy and practicality by continuously learning business data. NeMo microservices solve this problem by creating what Nvidia describes as "data flywheel", allowing AI systems to remain relevant through continuous exposure to enterprise information and user interaction. This newly launched toolkit contains five key microservices: NeMo Customizer handles fine-tuning of large language models with higher training throughput. NeMo Evaluator provides simplified evaluation of AI models for custom benchmarks. NeMo Guardrails implements security controls to maintain compliance and appropriateness
 AI Paints A New Picture For The Future Of Art And DesignApr 26, 2025 am 11:10 AM
AI Paints A New Picture For The Future Of Art And DesignApr 26, 2025 am 11:10 AMAI: The Future of Art and Design Artificial intelligence (AI) is changing the field of art and design in unprecedented ways, and its impact is no longer limited to amateurs, but more profoundly affecting professionals. Artwork and design schemes generated by AI are rapidly replacing traditional material images and designers in many transactional design activities such as advertising, social media image generation and web design. However, professional artists and designers also find the practical value of AI. They use AI as an auxiliary tool to explore new aesthetic possibilities, blend different styles, and create novel visual effects. AI helps artists and designers automate repetitive tasks, propose different design elements and provide creative input. AI supports style transfer, which is to apply a style of image
 How Zoom Is Revolutionizing Work With Agentic AI: From Meetings To MilestonesApr 26, 2025 am 11:09 AM
How Zoom Is Revolutionizing Work With Agentic AI: From Meetings To MilestonesApr 26, 2025 am 11:09 AMZoom, initially known for its video conferencing platform, is leading a workplace revolution with its innovative use of agentic AI. A recent conversation with Zoom's CTO, XD Huang, revealed the company's ambitious vision. Defining Agentic AI Huang d
 The Existential Threat To UniversitiesApr 26, 2025 am 11:08 AM
The Existential Threat To UniversitiesApr 26, 2025 am 11:08 AMWill AI revolutionize education? This question is prompting serious reflection among educators and stakeholders. The integration of AI into education presents both opportunities and challenges. As Matthew Lynch of The Tech Edvocate notes, universit
 The Prototype: American Scientists Are Looking For Jobs AbroadApr 26, 2025 am 11:07 AM
The Prototype: American Scientists Are Looking For Jobs AbroadApr 26, 2025 am 11:07 AMThe development of scientific research and technology in the United States may face challenges, perhaps due to budget cuts. According to Nature, the number of American scientists applying for overseas jobs increased by 32% from January to March 2025 compared with the same period in 2024. A previous poll showed that 75% of the researchers surveyed were considering searching for jobs in Europe and Canada. Hundreds of NIH and NSF grants have been terminated in the past few months, with NIH’s new grants down by about $2.3 billion this year, a drop of nearly one-third. The leaked budget proposal shows that the Trump administration is considering sharply cutting budgets for scientific institutions, with a possible reduction of up to 50%. The turmoil in the field of basic research has also affected one of the major advantages of the United States: attracting overseas talents. 35
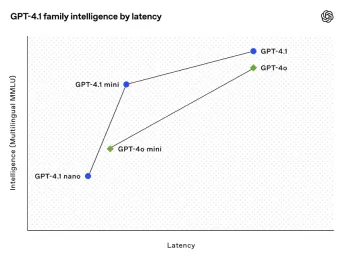 All About Open AI's Latest GPT 4.1 Family - Analytics VidhyaApr 26, 2025 am 10:19 AM
All About Open AI's Latest GPT 4.1 Family - Analytics VidhyaApr 26, 2025 am 10:19 AMOpenAI unveils the powerful GPT-4.1 series: a family of three advanced language models designed for real-world applications. This significant leap forward offers faster response times, enhanced comprehension, and drastically reduced costs compared t


Hot AI Tools

Undresser.AI Undress
AI-powered app for creating realistic nude photos

AI Clothes Remover
Online AI tool for removing clothes from photos.

Undress AI Tool
Undress images for free

Clothoff.io
AI clothes remover

Video Face Swap
Swap faces in any video effortlessly with our completely free AI face swap tool!

Hot Article

Hot Tools

Safe Exam Browser
Safe Exam Browser is a secure browser environment for taking online exams securely. This software turns any computer into a secure workstation. It controls access to any utility and prevents students from using unauthorized resources.

Dreamweaver Mac version
Visual web development tools

WebStorm Mac version
Useful JavaScript development tools

EditPlus Chinese cracked version
Small size, syntax highlighting, does not support code prompt function

mPDF
mPDF is a PHP library that can generate PDF files from UTF-8 encoded HTML. The original author, Ian Back, wrote mPDF to output PDF files "on the fly" from his website and handle different languages. It is slower than original scripts like HTML2FPDF and produces larger files when using Unicode fonts, but supports CSS styles etc. and has a lot of enhancements. Supports almost all languages, including RTL (Arabic and Hebrew) and CJK (Chinese, Japanese and Korean). Supports nested block-level elements (such as P, DIV),





