 Technology peripherals
Technology peripherals AI
AI The research team of the Chinese Academy of Sciences released two important papers: the release of the first large-scale model of the basis of life across species, and the release of a new AI model for cell fate prediction
The research team of the Chinese Academy of Sciences released two important papers: the release of the first large-scale model of the basis of life across species, and the release of a new AI model for cell fate prediction
Author | Chinese Academy of Sciences Multidisciplinary Research Team
Editor | ScienceAI
Known as one of the three major scientific projects of mankind in the 20th century The genome project has kicked off an in-depth analysis of the mysteries of life. Due to the multi-dimensional and highly dynamic nature of life processes, it is difficult for traditional experimental research methods to systematically and accurately decipher the underlying common laws of the genetic code. It is urgent to use powerful computing technology to achieve representation modeling and knowledge discovery of genetic data.
Currently, artificial intelligence technology with large models as the core has triggered revolutions in fields such as computer vision and natural language understanding, demonstrating in-depth understanding of data and knowledge, and is expected to be applied in the field of life science research, systems To accurately decipher the underlying common laws of genetic codes
Recently, the "Xcompass Consortium" (Xcompass Consortium) composed of a multi-disciplinary interdisciplinary research team of the Chinese Academy of Sciences has made important breakthroughs in artificial intelligence empowering life science research. Successfully Constructed the world's first large-scale model of the basis of cross-species life - GeneCompass. This model integrates the transcriptome data of more than 126 million single cells of humans and mice, and integrates four types of prior knowledge including promoter sequences and gene co-expression relationships. The number of basic model parameters reaches 130 million, realizing the control of gene expression. Panoramic learning and understanding of regulatory laws simultaneously supports prediction of cell state changes and accurate analysis of various life processes, demonstrating the great potential of artificial intelligence in empowering life science research.
The study is titled "GeneCompass: Deciphering Universal Gene Regulatory Mechanisms with Knowledge-Informed Cross-Species Foundation Model" and was published on bioRxiv.

Paper link: https://www.biorxiv.org/content/10.1101/2023.09.26.559542v1
In addition, The team also simultaneously released a gene regulatory network generation model based on transfer learning, CellPolaris, which can accurately identify core factors for cell fate conversion and has the ability to simulate transcription factor perturbations.
The research is titled "CellPolaris: Decoding Cell Fate through Generalization Transfer Learning of Gene Regulatory Networks" and was published on bioRxiv.

GeneCompass: The first large-scale model of the basis of life across species
Individual mammals typically contain tens of thousands to tens of trillions of cells. Although all cells in an individual contain the same genetic sequence, the fate and function of each cell vary widely due to its unique spatiotemporal context. Such a sophisticated life process is controlled by a complex gene expression regulation system
In order to enhance the understanding of the essential laws of life and innovate the diagnosis and treatment of various major diseases, it is necessary to study the ubiquitous gene regulation mechanisms of life. Explore deeper. However, traditional research methods have low throughput and are limited to a single model organism, and cannot reveal complex gene regulatory mechanisms. In recent years, breakthroughs in single-cell omics technology have produced a large number of gene expression profile data of different types of cells. , providing a data basis for interpreting gene-gene interactions. At the same time, the development of deep learning, especially the emergence of large generative models, can comprehensively summarize the nonlinear regulation mechanism of massive data learning in different cell states, bringing unprecedented opportunities to life science research.
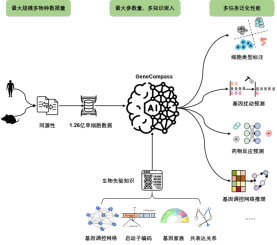
A large model of the basics of life across species, including 120 million cells and 130 million parametersCurrently, a single species has been obtained worldwide The scale of single-cell transcriptome data is only in the tens of millions, which is difficult to fully support the training of large models of basic life models used to analyze complex life processes.
The team collected open source single-cell transcriptome data from different species, and through pre-processing processes such as screening, cleaning, and normalization, established the largest known high-quality database, including more than 126 million cells in mice and humans. The training data set scCompass-126M adopts a deep learning architecture based on the Transformer self-attention mechanism, which can capture the long-term dynamic correlation between different genes in different cell backgrounds, and the model parameter size reaches 130 million. In order to achieve high-resolution characterization of life processes, GeneCompass dual-encodes gene numbers and expression levels for the first time, enabling effective and sensitive extraction of correlations between genes. This enables GeneCompass to provide more precise analysis of gene-gene interactions under a variety of specific conditions, such as cell types and perturbation states.
Embedding prior knowledge during pre-training can effectively improve model performance
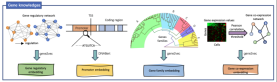
The model effectively integrates promoter sequences, known gene regulatory networks, gene family information and gene co- Expressing the relationship between four kinds of biological prior knowledge, adding human annotation information encoding, improves the understanding of complex feature correlations between biological data. Through training and integrating data information and prior knowledge of different species, GeneCompass is expected to improve the efficiency and accuracy of traditional biological research and bring new entry points to complex life science problems that cannot yet be broken through.
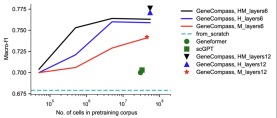
Scale effect prompts model training to capture the conservative laws of biological evolution
The team found that models pre-trained on large-scale cross-species data can perform better on single-species subtasks It is consistent with the scaling law: larger-scale multi-species pre-training data can produce better pre-training representations and further improve the performance of downstream tasks. This finding shows that there are conserved gene regulation patterns between species, and that these patterns can be learned and understood by pre-trained models. At the same time, this also means that with the expansion of species and data, model performance is expected to continue to improve
Multi-task performance advantages show the powerful generalization ability of the basic large model
As the largest cross-species pre-trained basic life model with knowledge embedding to date, GeneCompass can realize multiple cross-species Transfer learning for downstream tasks, and achieving better performance than existing methods in cell type annotation, quantitative gene perturbation prediction, drug sensitivity analysis, etc. This fully demonstrates the strategic advantages of pre-training based on multi-species unlabeled big data and then using different sub-task data for model fine-tuning. It is expected to become a universal solution for analyzing and predicting various biological problems related to gene-cell characteristics.
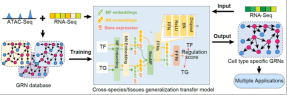
Cell polarization: Transfer learning decodes gene regulatory networks and predicts cell fate changes
Using transfer learning to generate cells Specific gene regulatory network
The team also developed a set of gene regulatory network construction AI models based on generalized transfer learning, called CellPolaris. The model first sorts out hundreds of sets of transcriptome and chromatin accessibility data in matching cell scenarios to build a high-quality gene regulatory network, and then uses the generalized transfer learning model to generate more genes in cell scenarios using only transcriptome data. regulatory network. Then, using the generated high-confidence gene regulatory network, we developed a tool for identifying core transcription factors in cell fate transitions and a transcription factor perturbation simulation tool based on a probabilistic graphical model. This model can effectively identify the core factors of cell fate conversion and realize the simulation of transcription factor perturbation. It has important application value in the analysis of gene regulatory mechanisms and the discovery of disease-causing genes.
× [Biological Neuroscience Mathematics Physics Chemistry Materials]
The above is the detailed content of The research team of the Chinese Academy of Sciences released two important papers: the release of the first large-scale model of the basis of life across species, and the release of a new AI model for cell fate prediction. For more information, please follow other related articles on the PHP Chinese website!
 The AI Skills Gap Is Slowing Down Supply ChainsApr 26, 2025 am 11:13 AM
The AI Skills Gap Is Slowing Down Supply ChainsApr 26, 2025 am 11:13 AMThe term "AI-ready workforce" is frequently used, but what does it truly mean in the supply chain industry? According to Abe Eshkenazi, CEO of the Association for Supply Chain Management (ASCM), it signifies professionals capable of critic
 How One Company Is Quietly Working To Transform AI ForeverApr 26, 2025 am 11:12 AM
How One Company Is Quietly Working To Transform AI ForeverApr 26, 2025 am 11:12 AMThe decentralized AI revolution is quietly gaining momentum. This Friday in Austin, Texas, the Bittensor Endgame Summit marks a pivotal moment, transitioning decentralized AI (DeAI) from theory to practical application. Unlike the glitzy commercial
 Nvidia Releases NeMo Microservices To Streamline AI Agent DevelopmentApr 26, 2025 am 11:11 AM
Nvidia Releases NeMo Microservices To Streamline AI Agent DevelopmentApr 26, 2025 am 11:11 AMEnterprise AI faces data integration challenges The application of enterprise AI faces a major challenge: building systems that can maintain accuracy and practicality by continuously learning business data. NeMo microservices solve this problem by creating what Nvidia describes as "data flywheel", allowing AI systems to remain relevant through continuous exposure to enterprise information and user interaction. This newly launched toolkit contains five key microservices: NeMo Customizer handles fine-tuning of large language models with higher training throughput. NeMo Evaluator provides simplified evaluation of AI models for custom benchmarks. NeMo Guardrails implements security controls to maintain compliance and appropriateness
 AI Paints A New Picture For The Future Of Art And DesignApr 26, 2025 am 11:10 AM
AI Paints A New Picture For The Future Of Art And DesignApr 26, 2025 am 11:10 AMAI: The Future of Art and Design Artificial intelligence (AI) is changing the field of art and design in unprecedented ways, and its impact is no longer limited to amateurs, but more profoundly affecting professionals. Artwork and design schemes generated by AI are rapidly replacing traditional material images and designers in many transactional design activities such as advertising, social media image generation and web design. However, professional artists and designers also find the practical value of AI. They use AI as an auxiliary tool to explore new aesthetic possibilities, blend different styles, and create novel visual effects. AI helps artists and designers automate repetitive tasks, propose different design elements and provide creative input. AI supports style transfer, which is to apply a style of image
 How Zoom Is Revolutionizing Work With Agentic AI: From Meetings To MilestonesApr 26, 2025 am 11:09 AM
How Zoom Is Revolutionizing Work With Agentic AI: From Meetings To MilestonesApr 26, 2025 am 11:09 AMZoom, initially known for its video conferencing platform, is leading a workplace revolution with its innovative use of agentic AI. A recent conversation with Zoom's CTO, XD Huang, revealed the company's ambitious vision. Defining Agentic AI Huang d
 The Existential Threat To UniversitiesApr 26, 2025 am 11:08 AM
The Existential Threat To UniversitiesApr 26, 2025 am 11:08 AMWill AI revolutionize education? This question is prompting serious reflection among educators and stakeholders. The integration of AI into education presents both opportunities and challenges. As Matthew Lynch of The Tech Edvocate notes, universit
 The Prototype: American Scientists Are Looking For Jobs AbroadApr 26, 2025 am 11:07 AM
The Prototype: American Scientists Are Looking For Jobs AbroadApr 26, 2025 am 11:07 AMThe development of scientific research and technology in the United States may face challenges, perhaps due to budget cuts. According to Nature, the number of American scientists applying for overseas jobs increased by 32% from January to March 2025 compared with the same period in 2024. A previous poll showed that 75% of the researchers surveyed were considering searching for jobs in Europe and Canada. Hundreds of NIH and NSF grants have been terminated in the past few months, with NIH’s new grants down by about $2.3 billion this year, a drop of nearly one-third. The leaked budget proposal shows that the Trump administration is considering sharply cutting budgets for scientific institutions, with a possible reduction of up to 50%. The turmoil in the field of basic research has also affected one of the major advantages of the United States: attracting overseas talents. 35
 All About Open AI's Latest GPT 4.1 Family - Analytics VidhyaApr 26, 2025 am 10:19 AM
All About Open AI's Latest GPT 4.1 Family - Analytics VidhyaApr 26, 2025 am 10:19 AMOpenAI unveils the powerful GPT-4.1 series: a family of three advanced language models designed for real-world applications. This significant leap forward offers faster response times, enhanced comprehension, and drastically reduced costs compared t


Hot AI Tools

Undresser.AI Undress
AI-powered app for creating realistic nude photos

AI Clothes Remover
Online AI tool for removing clothes from photos.

Undress AI Tool
Undress images for free

Clothoff.io
AI clothes remover

Video Face Swap
Swap faces in any video effortlessly with our completely free AI face swap tool!

Hot Article

Hot Tools

Atom editor mac version download
The most popular open source editor

MantisBT
Mantis is an easy-to-deploy web-based defect tracking tool designed to aid in product defect tracking. It requires PHP, MySQL and a web server. Check out our demo and hosting services.

DVWA
Damn Vulnerable Web App (DVWA) is a PHP/MySQL web application that is very vulnerable. Its main goals are to be an aid for security professionals to test their skills and tools in a legal environment, to help web developers better understand the process of securing web applications, and to help teachers/students teach/learn in a classroom environment Web application security. The goal of DVWA is to practice some of the most common web vulnerabilities through a simple and straightforward interface, with varying degrees of difficulty. Please note that this software

WebStorm Mac version
Useful JavaScript development tools

SAP NetWeaver Server Adapter for Eclipse
Integrate Eclipse with SAP NetWeaver application server.





