 Technology peripherals
Technology peripherals AI
AI Google Scientist Nature comments: How artificial intelligence can better understand the brain
Google Scientist Nature comments: How artificial intelligence can better understand the brainGoogle Scientist Nature comments: How artificial intelligence can better understand the brain

Compilation | Green Dior
On November 7, 2023, Viren Jain, senior research scientist at Google Research and head of connectomics of the Google team, published in "Nature" A review article titled "How AI could lead to a better understanding of the brain".
Paper link: https://www.nature.com/articles/d41586-023-03426-3
Can computers be programmed to simulate the brain? ? It's a question that mathematicians, theorists and experimentalists have long asked - whether out of a desire to create artificial intelligence (AI) or because its behavior can only be understood if mathematics or computers can reproduce it Complex systems like the brain. To try to answer this question, researchers have been developing simplified models of the brain's neural networks since the 1940s. In fact, today’s explosion of machine learning can be traced back to early work inspired by biological systems.
However, the results of these efforts now allow researchers to ask a slightly different question: Can machine learning be used to build computational models that simulate brain activity?
At the heart of these developments is increasing amounts of brain data. Starting in the 1970s, neuroscientists have been producing connectomes, maps of neuronal connections and morphology that capture static representations of the brain at a given moment, and this research has since intensified. In addition to these advances, researchers have also improved their ability to make functional recordings that can measure changes in neural activity over time at the resolution of single cells. Meanwhile, the field of transcriptomics allows researchers to measure gene activity in tissue samples and even map when and where that activity occurs.
To date, few attempts have been made to connect these different data sources or to collect them simultaneously from the entire brain of the same sample. But as the level of detail, size, and number of data sets increase, especially for the brains of relatively simple model organisms, machine learning systems are making a new approach to brain modeling feasible. This involves training artificial intelligence programs on connectome and other data to reproduce the neural activity you would expect to find in biological systems.
Computational neuroscientists and others need to solve some challenges before they can begin using machine learning to build simulations of the entire brain. However, a hybrid approach that combines information from traditional brain modeling techniques with machine learning systems trained on different data sets can make the entire effort more rigorous and informative.
Brain Mapping
The quest to map the brain began nearly half a century ago with 15 years of painstaking research in the nematode Caenorhabditis elegans. Over the past two decades, developments in automated tissue sectioning and imaging have made anatomical data more accessible to researchers, while advances in computing and automated image analysis have transformed the analysis of these data sets.
Connectomes have now been generated for the entire brain of C. elegans, larval and adult Drosophila melanogaster, and for small portions (one-thousandth and one-millionth, respectively) of mouse and human brains.
The anatomy diagrams produced so far contain major flaws. Imaging methods have not been able to map electrical connections at scale along with chemical synaptic connections. Researchers have focused primarily on neurons, although the non-neuronal glial cells that provide support to neurons appear to play a crucial role in the flow of information in the nervous system. Much is still unknown about the genes expressed and the proteins present in the neurons and other cells that were mapped.
Still, such maps have yielded some insights. In Drosophila melanogaster, for example, connectomics allows researchers to identify the mechanisms behind neural circuits responsible for behaviors such as aggression. The brain map also revealed how fruit flies compute information in the circuits responsible for knowing where they are and how to get from one place to another. In zebrafish (Danio rerio) larvae, connectomics helped reveal the workings of synaptic circuits underlying odor classification, control of eye position and movement, and navigation.
Efforts that could eventually generate the entire mouse brain connectome are underway—although with current methods, this could take a decade or more. The mouse brain is nearly 1,000 times larger than the brain of Drosophila melanogaster, which is composed of about 150,000 neurons.
In addition to all these advances in connectomics, researchers are leveraging single-cell and spatial transcriptomics to capture gene expression patterns with ever-increasing accuracy and specificity. Various techniques also allow researchers to record neural activity from the entire brain of a vertebrate animal for hours at a time. In the case of the larval zebrafish brain, this means recording from nearly 100,000 neurons. These include proteins with fluorescent properties that change in response to changes in voltage or calcium levels, and microscopy techniques that enable 3D imaging of living brains at single-cell resolution. (Recordings of neural activity in this way provide a less accurate picture than electrophysiological recordings, but much better than non-invasive methods such as functional magnetic resonance imaging.)
Mathematics and Physics
When trying to simulate brain activity patterns, scientists mainly use physics-based methods. This requires generating a simulation of a nervous system or parts of a nervous system using a mathematical description of the behavior of real neurons or parts of a real nervous system. It also requires making informed guesses about aspects of the circuit that have not been verified by observation, such as network connectivity.
In some cases, speculation is extensive (see "Mystery Model") but in other ways, anatomical maps at single-cell and single-synapse resolution help researchers refute and generate hypotheses.
Mysterious Model
Due to a lack of data, it is difficult to evaluate whether certain neural network models capture what happens in real systems.
The controversial European Human Brain Project, which ended in September, originally aimed to computationally simulate the entire human brain. Although that goal was abandoned, the project did simulate parts of rodent and human brains, including tens of thousands of neurons in a rodent hippocampus model, based on limited biological measurements and a variety of synthetic data-generating procedures.
A major problem with this approach is that in the absence of detailed anatomical or functional diagrams, it is difficult to assess how accurately the resulting simulation captures what is happening in the biological system.
For about seventy years, neuroscientists have been refining theoretical descriptions of the circuits that enable the calculation of movement in Drosophila melanogaster. Since its completion in 2013, the motion detection circuit connectome, and subsequently the larger flight connectome, has provided detailed circuit diagrams that support some hypotheses about how the circuit works.
However, data collected from real neural networks also highlights the limitations of anatomy-driven approaches.
For example, a neural circuit model completed in the 1990s included a detailed analysis of the connectivity and physiology of the approximately 30 neurons that make up the crab (Cancer borealis) orogastric ganglion (which controls the animal's stomach). structure of movement). By measuring the activity of neurons under various conditions, the researchers found that even for relatively small collections of neurons, seemingly subtle changes, such as the introduction of a neuromodulator (a substance that changes the properties of neurons and synapses), , will also completely change the behavior of the circuit. This suggests that even with connectomes and other rich data sets to guide and constrain hypotheses about neural circuits, today's data may not be detailed enough for modelers to capture what is happening in biological systems.
This is an area where machine learning can provide a way forward.
By optimizing thousands or even billions of parameters guided by connectome and other data, machine learning models can be trained to produce neural network behavior that is consistent with real neural network behavior - using cellular resolution capabilities Record the measurement.
This machine learning model can incorporate information from traditional brain modeling techniques, such as the Hodgkin-Huxley model, which describes action potentials in neurons (across how changes in membrane voltage) are initiated and propagated using optimized parametric connectivity maps, functional activity recordings, or other data sets obtained for the whole brain. Alternatively, machine learning models can contain “black box” architectures that contain little explicitly specified biological knowledge but contain billions or hundreds of billions of parameters, all of which are empirically optimized.
For example, researchers can evaluate such models by comparing predictions of the system's neural activity to recordings of actual biological systems. Crucially, when machine learning programs are given untrained data, they evaluate how the model's predictions compare—as is standard practice when evaluating machine learning systems.
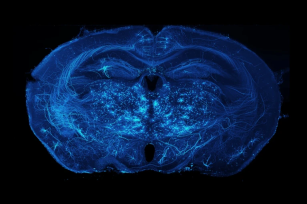
Axonal projections of neurons in the mouse brain. (Source: Adam Glaser, Jayaram Chandrashekar, Karel Svoboda, Allen Institute for Neurodynamics)
This approach will enable more rigorous modeling of brains containing thousands or more neurons. For example, researchers will be able to evaluate whether simpler models that are easier to compute simulate neural networks better than more complex models that provide more detailed biophysical information, and vice versa.
Machine learning is already being used in this way to improve understanding of other extremely complex systems. For example, since the 1950s, weather prediction systems have typically relied on carefully constructed mathematical models of meteorological phenomena, and modern systems are the result of the iterative refinement of such models by hundreds of researchers. However, over the past five years or so, researchers have developed several weather prediction systems that leverage machine learning. For example, these contain fewer assumptions related to how pressure gradients drive changes in wind speed and how wind speed moves moisture through the atmosphere. Instead, millions of parameters are optimized through machine learning to produce simulated weather behavior that is consistent with a database of past weather patterns.
This way of doing things does come with some challenges. Even if a model makes accurate predictions, it's difficult to explain how it does it. Additionally, models often fail to predict scenarios that are not included in their training data. A weather model trained to predict the next few days has difficulty extrapolating forecasts to weeks or months into the future. But in some cases - for forecasting rainfall several hours into the future - machine learning methods have outperformed traditional methods. Machine learning models also have practical advantages. They use simpler underlying code and can be used by scientists with less specialized meteorological knowledge.
For brain modeling, on the one hand, this approach could help fill some of the gaps in current datasets and reduce the need for more detailed measurements of individual biological components, such as individual neurons. On the other hand, as more comprehensive data sets become available, incorporating the data into the model will become simple.
Think Big
In order to realize this idea, some challenges need to be solved.
Machine learning programs are only as good as the data used to train and evaluate them. Therefore, neuroscientists should aim to obtain data sets from the entire brain of a sample—or even the entire body, if this becomes more feasible. Although it is easier to collect data from certain parts of the brain, using machine learning to model highly interconnected systems, such as neural networks, is unlikely to yield useful information if many parts of the system are not present in the underlying data. .
Researchers should also work to obtain anatomical maps of neural connections and functional recordings (and perhaps in the future gene expression maps) from whole brains from the same sample. Currently, either group tends to focus solely on getting one or the other, rather than both.
With only 302 neurons, the C. elegans nervous system may have enough hardwiring to allow researchers to assume that the connectivity map obtained from one sample will be the same for any other sample - although some studies Shows otherwise. But for larger nervous systems, such as those of Drosophila melanogaster and zebrafish larvae, the connectome variation between samples is significant, so brain models should be trained on structural and functional data obtained from the same sample.
Currently, this can only be achieved in two common model organisms. The bodies of C. elegans and zebrafish larvae are transparent, meaning researchers can make functional recordings from the organism's entire brain and pinpoint the activity of individual neurons. Following such recordings, the animals can be killed immediately, embedded in resin and sectioned, and anatomical measurements of neural connections made. In the future, however, researchers could expand the range of organisms for which such combined data acquisition is possible, for example, by developing new non-invasive methods, possibly using ultrasound, to record neural activity at high resolution.
Obtaining such multimodal datasets in the same sample requires extensive collaboration among researchers, investment in large team science, and increased funding agency support for a more comprehensive effort. But there is precedent for this approach, such as the U.S. Intelligence Advanced Research Program Activity's MICrONS project, which obtained functional and anatomical data on 1 cubic millimeter of mouse brain between 2016 and 2021.
In addition to obtaining this data, neuroscientists need to agree on key modeling goals and quantitative metrics to measure progress. Should the goal of the model be to predict the behavior of individual neurons based on past states or the entire brain? Should the activity of a single neuron be the key metric, or should it be the percentage of hundreds of thousands of active neurons? Likewise, what constitutes an accurate representation of neural activity in a biological system? Formal, agreed-upon benchmarks are essential for comparing modeling approaches and tracking progress over time.
Finally, to present brain modeling challenges to diverse communities, including computational neuroscientists and machine learning experts, researchers need to clarify to the broader scientific community which modeling tasks are the highest priority and which metrics should be used to evaluate the performance of the model. WeatherBench, an online platform that provides a framework for evaluating and comparing weather forecast models, provides a useful template.
Complexity of Key Technologies
Some will question—rightly so—whether machine learning approaches to brain modeling are scientifically useful. Could the problem of trying to understand how the brain work simply be replaced by the problem of trying to understand how large artificial networks work?
However, it is encouraging to use similar methods in a branch of neuroscience involved in determining how the brain processes and encodes sensory stimuli, such as sight and smell. Researchers are increasingly using classically modeled neural networks, in which some biological details are specified, combined with machine learning systems. The latter are trained on large visual or audio data sets to reproduce the visual or auditory abilities of the neural system, such as image recognition. The resulting network showed striking similarities to biological networks, but was easier to analyze and interrogate than true neural networks.
For now, perhaps it is enough to ask whether data from current brain atlases and other work can train machine learning models to reproduce neural activity that corresponds to what is seen in biological systems. Here, even failure can be fun - suggesting that mapping research must go deeper.
The above is the detailed content of Google Scientist Nature comments: How artificial intelligence can better understand the brain. For more information, please follow other related articles on the PHP Chinese website!
 Personal Hacking Will Be A Pretty Fierce BearMay 11, 2025 am 11:09 AM
Personal Hacking Will Be A Pretty Fierce BearMay 11, 2025 am 11:09 AMCyberattacks are evolving. Gone are the days of generic phishing emails. The future of cybercrime is hyper-personalized, leveraging readily available online data and AI to craft highly targeted attacks. Imagine a scammer who knows your job, your f
 Pope Leo XIV Reveals How AI Influenced His Name ChoiceMay 11, 2025 am 11:07 AM
Pope Leo XIV Reveals How AI Influenced His Name ChoiceMay 11, 2025 am 11:07 AMIn his inaugural address to the College of Cardinals, Chicago-born Robert Francis Prevost, the newly elected Pope Leo XIV, discussed the influence of his namesake, Pope Leo XIII, whose papacy (1878-1903) coincided with the dawn of the automobile and
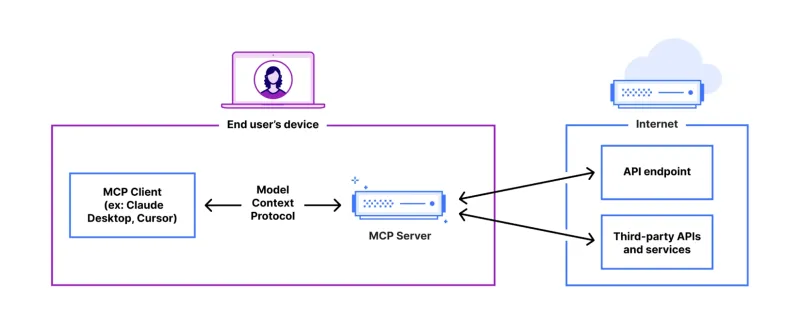 FastAPI-MCP Tutorial for Beginners and Experts - Analytics VidhyaMay 11, 2025 am 10:56 AM
FastAPI-MCP Tutorial for Beginners and Experts - Analytics VidhyaMay 11, 2025 am 10:56 AMThis tutorial demonstrates how to integrate your Large Language Model (LLM) with external tools using the Model Context Protocol (MCP) and FastAPI. We'll build a simple web application using FastAPI and convert it into an MCP server, enabling your L
 Dia-1.6B TTS : Best Text-to-Dialogue Generation Model - Analytics VidhyaMay 11, 2025 am 10:27 AM
Dia-1.6B TTS : Best Text-to-Dialogue Generation Model - Analytics VidhyaMay 11, 2025 am 10:27 AMExplore Dia-1.6B: A groundbreaking text-to-speech model developed by two undergraduates with zero funding! This 1.6 billion parameter model generates remarkably realistic speech, including nonverbal cues like laughter and sneezes. This article guide
 3 Ways AI Can Make Mentorship More Meaningful Than EverMay 10, 2025 am 11:17 AM
3 Ways AI Can Make Mentorship More Meaningful Than EverMay 10, 2025 am 11:17 AMI wholeheartedly agree. My success is inextricably linked to the guidance of my mentors. Their insights, particularly regarding business management, formed the bedrock of my beliefs and practices. This experience underscores my commitment to mentor
 AI Unearths New Potential In The Mining IndustryMay 10, 2025 am 11:16 AM
AI Unearths New Potential In The Mining IndustryMay 10, 2025 am 11:16 AMAI Enhanced Mining Equipment The mining operation environment is harsh and dangerous. Artificial intelligence systems help improve overall efficiency and security by removing humans from the most dangerous environments and enhancing human capabilities. Artificial intelligence is increasingly used to power autonomous trucks, drills and loaders used in mining operations. These AI-powered vehicles can operate accurately in hazardous environments, thereby increasing safety and productivity. Some companies have developed autonomous mining vehicles for large-scale mining operations. Equipment operating in challenging environments requires ongoing maintenance. However, maintenance can keep critical devices offline and consume resources. More precise maintenance means increased uptime for expensive and necessary equipment and significant cost savings. AI-driven
 Why AI Agents Will Trigger The Biggest Workplace Revolution In 25 YearsMay 10, 2025 am 11:15 AM
Why AI Agents Will Trigger The Biggest Workplace Revolution In 25 YearsMay 10, 2025 am 11:15 AMMarc Benioff, Salesforce CEO, predicts a monumental workplace revolution driven by AI agents, a transformation already underway within Salesforce and its client base. He envisions a shift from traditional markets to a vastly larger market focused on
 AI HR Is Going To Rock Our Worlds As AI Adoption SoarsMay 10, 2025 am 11:14 AM
AI HR Is Going To Rock Our Worlds As AI Adoption SoarsMay 10, 2025 am 11:14 AMThe Rise of AI in HR: Navigating a Workforce with Robot Colleagues The integration of AI into human resources (HR) is no longer a futuristic concept; it's rapidly becoming the new reality. This shift impacts both HR professionals and employees, dem


Hot AI Tools

Undresser.AI Undress
AI-powered app for creating realistic nude photos

AI Clothes Remover
Online AI tool for removing clothes from photos.

Undress AI Tool
Undress images for free

Clothoff.io
AI clothes remover

Video Face Swap
Swap faces in any video effortlessly with our completely free AI face swap tool!

Hot Article

Hot Tools

Notepad++7.3.1
Easy-to-use and free code editor

SublimeText3 Chinese version
Chinese version, very easy to use

Zend Studio 13.0.1
Powerful PHP integrated development environment

SublimeText3 Linux new version
SublimeText3 Linux latest version

WebStorm Mac version
Useful JavaScript development tools






