 Technology peripherals
Technology peripherals AI
AI Open source! Hong Kong Chinese, MIT, and Fudan propose the first RNA cornerstone model
Open source! Hong Kong Chinese, MIT, and Fudan propose the first RNA cornerstone modelOpen source! Hong Kong Chinese, MIT, and Fudan propose the first RNA cornerstone model
Unlike the protein field, research in the RNA field often lacks sufficient annotation data. For example, 3D data only has more than 1,000 RNAs. This greatly limits the development of machine learning methods in RNA structure-function prediction tasks.
In order to make up for the lack of annotated data, this article demonstrates a cornerstone model that can provide rich structural and functional knowledge for various RNA studies - RNA foundation model ( RNA-FM). As the world's first RNA cornerstone model trained in an unsupervised manner based on 23 million unlabeled RNA sequences, RNA-FM mines the evolutionary and structural patterns contained in RNA sequences.
It is worth noting that RNA-FM only needs to match a simple downstream model or only provide embedding, and it can achieve performance far exceeding SOTA in many downstream tasks, such as It can be improved by 20% in secondary structure prediction and 30% in distance map prediction. Large-scale experiments have proven that the model is highly generalizable and can even be used for COVID-19 and regulatory fragments of mRNA.
- ##Preprint of the paper: https://arxiv. org/abs/2204.00300
- Code and model: https://github.com/ml4bio/RNA-FM
- Server: https://proj.cse.cuhk.edu.hk/rnafm
In recent years, biological computing methods based on deep learning have made breakthrough progress in the field of proteins. The most famous milestone is the end-to-end protein 3D structure prediction framework AlphaFold2 developed by the Google DeepMind team. However, protein is only one type of many biological molecules. Gene (DNA/RNA), as the source of protein production, contains more basic information than the latter and has more important research value.
Generally speaking, proteins are the products of translation from RNA used for coding, that is, mRNA. A fixed mRNA can be translated into a fixed protein sequence. In fact, this part of coding RNA only accounts for 2% of all RNA sequences, and the remaining 98% is non-coding RNA (ncRNA). Although ncRNAs are not directly "translated" into proteins, they fold into tertiary structures with specific functions and play a regulatory role in the translation process of mRNA or other biological functions. Therefore, analyzing the structure and function of ncRNA is a more basic and complex research than protein analysis.
However, compared to the protein field, where computational methods are more mature, RNA-based structure and function prediction is still in its early stages, and computational methods originally applicable to the protein field are difficult to directly migrate. to the RNA field. The main limitation of these computational methods is that annotation of RNA data is usually difficult to obtain, and it requires a lot of experimental resources and time to complete the annotation of a small amount of data. Most computational methods require a large amount of annotated data for supervision to achieve high performance. Although there is not much annotated data, the RNA field has actually accumulated a lot of unannotated sequence data. The method of this article is to use these unlabeled data to provide additional effective information for various downstream tasks.
Based on this consideration, Hong Kong Chinese, MIT, Fudan and Shanghai Artificial Intelligence Laboratory teams proposed an unsupervised method to The RNA foundation model (RNA-FM) is trained on 23 million label-free pure RNA sequences. Although the data does not provide annotation information during the training process, RNA-FM still mines the evolutionary and structural patterns contained in these RNA sequences in an unsupervised manner.
If RNA-FM can be effectively applied to downstream RNA structure and function prediction tasks, these computational methods will surely benefit from the knowledge induced by RNA-FM and achieve better performance Performance improvements. The upstream pre-training and downstream migration and application framework of RNA-FM are shown in the figure below.
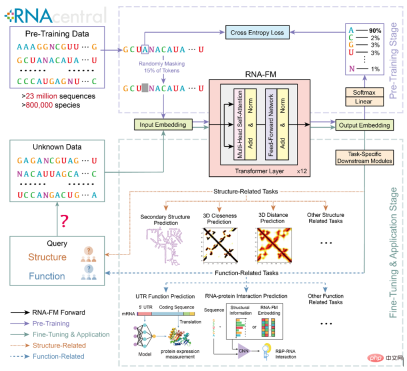
In order to confirm whether the pre-trained RNA-FM has learned "knowledge" from a large amount of unlabeled data and What kind of "knowledge" has been learned? The articleconducts a series of analyzes on embedding.
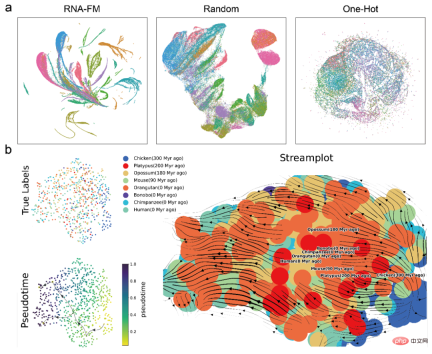
First, a simple clustering comparison of various features was conducted directly through UMAP, and it was found that the embedding from pre-trained RNA-FM was better than other Embedding forms clusters with more distinct RNA species. This means that the embedding of RNA-FM does contain structural or functional information for distinguishing RNA species.
Then, the article also uses trajectory inference (Trajectory inference) to predict the evolution of lncRNA from different species through RNA-FM embedding. From the streamplot below, the predicted pseudo-time of evolution between species is roughly consistent with the real species evolution information, indicating that RNA-FM embedding also contains part of the evolutionary information.
It is worth noting that, whether it is community information of RNA species or evolutionary information of lncRNA, RNA-FM has not been directly exposed to these labels during training. RNA-FM discovers patterns related to structure, function and evolution from pure sequences in a completely self-supervised manner.
More experimental results
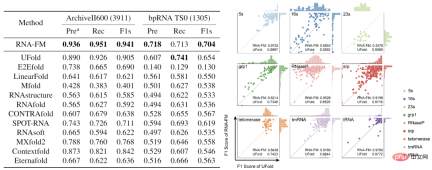
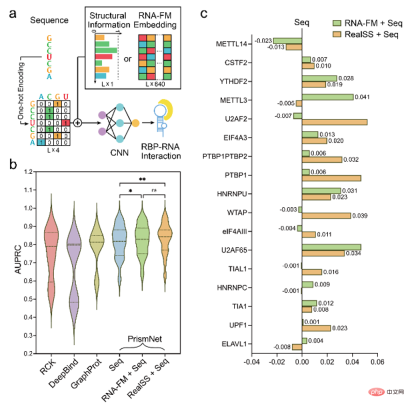
In addition to directly analyzing the embedding of RNA-FM , the article also attempts to introduce RNA-FM to various downstream RNA structure prediction tasks, including secondary structure, contact prediction, distance prediction, and tertiary structure prediction, and has achieved obvious results. promote.
Especially in terms of secondary structure prediction, the article uses RNA-FM as the backbone and only uses a simple ResNet network as the downstream model, surpassing two public data sets. The other 12 state-of-the-art methods are superior to the best UFold by 3-5 percentage points in F1score. In the head-to-head comparison with UFold, RNA-FM performs better in most RNA categories. More than UFold. If RNA-FM is combined with E2Efold, a further 5% performance improvement can be achieved.
Using RNA-FM to conduct a complete analysis of COVID-19 data, including using RNA-FM to accurately predict key regulatory elements in the COVID-19 reference genome (29870 nt), and using RNA-FM embedding to roughly predict the evolutionary trends of major COVID-19 variants. 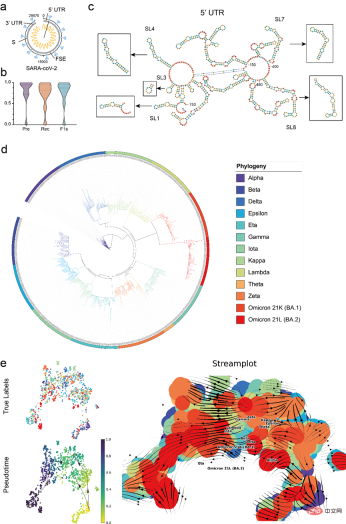
Therefore, the article
further attempts to introduce RNA-FM into downstream RNA function prediction tasks, such as using RNA-FM embedding. Prediction of RNA-protein roles.
Experiments have proven that the introduction of RNA-FM embedding improves the performance of the model, and in some cases even achieves prediction results that match real secondary structure information as input.
finally attempts to use RNA -FM performs functional prediction of protein expression based on 5'UTR on mRNA. Although mRNA does not belong to ncRNA, the 5'UTR on it is a region that is not translated but has regulatory functions, which is consistent with the characteristics of ncRNA and does not appear in the training data.
As you can see from the figure below, models that include RNA-FM embedding are always better than models that do not. Although the performance improvement is relatively limited, it partly shows that RNA-FM also has certain generalization ability on non-ncRNA data.
Conclusion
In general, this article uses unlabeled RNA sequence data to pre-train the language model RNA-FM, and through direct or indirect methods, a series of structural or functional Comprehensive verification on different tasks proves that RNA-FM can indeed effectively improve the performance of computing methods in downstream tasks.
The emergence of RNA-FM has alleviated the current situation of RNA labeled data to a certain extent, and provides other researchers with a convenient interface to access large quantities of unlabeled data, which will As a basic model in the RNA field, it provides strong support and help for various research in this field.
About the author
This article has two co-first authors. Chen Jiayang is a research assistant at the Chinese University of Hong Kong. Hu Zhihang is a doctoral candidate at the Chinese University of Hong Kong.
This article has two corresponding authors. Sun Siqi, young researcher at Fudan University Intelligent Complex Systems Laboratory and Shanghai Artificial Intelligence Laboratory, homepage https://intersun.github.io.
Li Yu, Assistant Professor at the Chinese University of Hong Kong, Visiting Assistant Professor at MIT James Collins Lab, Research Scientist at Broad Institute of MIT and Harvard, Visiting Scholar at Wyss Institute at Harvard University, Forbes 30 Under 30 Asia list–Class of 2022, Healthcare & Science. Home page: https://liyu95.com.
The above is the detailed content of Open source! Hong Kong Chinese, MIT, and Fudan propose the first RNA cornerstone model. For more information, please follow other related articles on the PHP Chinese website!
 From Friction To Flow: How AI Is Reshaping Legal WorkMay 09, 2025 am 11:29 AM
From Friction To Flow: How AI Is Reshaping Legal WorkMay 09, 2025 am 11:29 AMThe legal tech revolution is gaining momentum, pushing legal professionals to actively embrace AI solutions. Passive resistance is no longer a viable option for those aiming to stay competitive. Why is Technology Adoption Crucial? Legal professional
 This Is What AI Thinks Of You And Knows About YouMay 09, 2025 am 11:24 AM
This Is What AI Thinks Of You And Knows About YouMay 09, 2025 am 11:24 AMMany assume interactions with AI are anonymous, a stark contrast to human communication. However, AI actively profiles users during every chat. Every prompt, every word, is analyzed and categorized. Let's explore this critical aspect of the AI revo
 7 Steps To Building A Thriving, AI-Ready Corporate CultureMay 09, 2025 am 11:23 AM
7 Steps To Building A Thriving, AI-Ready Corporate CultureMay 09, 2025 am 11:23 AMA successful artificial intelligence strategy cannot be separated from strong corporate culture support. As Peter Drucker said, business operations depend on people, and so does the success of artificial intelligence. For organizations that actively embrace artificial intelligence, building a corporate culture that adapts to AI is crucial, and it even determines the success or failure of AI strategies. West Monroe recently released a practical guide to building a thriving AI-friendly corporate culture, and here are some key points: 1. Clarify the success model of AI: First of all, we must have a clear vision of how AI can empower business. An ideal AI operation culture can achieve a natural integration of work processes between humans and AI systems. AI is good at certain tasks, while humans are good at creativity and judgment
 Netflix New Scroll, Meta AI's Game Changers, Neuralink Valued At $8.5 BillionMay 09, 2025 am 11:22 AM
Netflix New Scroll, Meta AI's Game Changers, Neuralink Valued At $8.5 BillionMay 09, 2025 am 11:22 AMMeta upgrades AI assistant application, and the era of wearable AI is coming! The app, designed to compete with ChatGPT, offers standard AI features such as text, voice interaction, image generation and web search, but has now added geolocation capabilities for the first time. This means that Meta AI knows where you are and what you are viewing when answering your question. It uses your interests, location, profile and activity information to provide the latest situational information that was not possible before. The app also supports real-time translation, which completely changed the AI experience on Ray-Ban glasses and greatly improved its usefulness. The imposition of tariffs on foreign films is a naked exercise of power over the media and culture. If implemented, this will accelerate toward AI and virtual production
 Take These Steps Today To Protect Yourself Against AI CybercrimeMay 09, 2025 am 11:19 AM
Take These Steps Today To Protect Yourself Against AI CybercrimeMay 09, 2025 am 11:19 AMArtificial intelligence is revolutionizing the field of cybercrime, which forces us to learn new defensive skills. Cyber criminals are increasingly using powerful artificial intelligence technologies such as deep forgery and intelligent cyberattacks to fraud and destruction at an unprecedented scale. It is reported that 87% of global businesses have been targeted for AI cybercrime over the past year. So, how can we avoid becoming victims of this wave of smart crimes? Let’s explore how to identify risks and take protective measures at the individual and organizational level. How cybercriminals use artificial intelligence As technology advances, criminals are constantly looking for new ways to attack individuals, businesses and governments. The widespread use of artificial intelligence may be the latest aspect, but its potential harm is unprecedented. In particular, artificial intelligence
 A Symbiotic Dance: Navigating Loops Of Artificial And Natural PerceptionMay 09, 2025 am 11:13 AM
A Symbiotic Dance: Navigating Loops Of Artificial And Natural PerceptionMay 09, 2025 am 11:13 AMThe intricate relationship between artificial intelligence (AI) and human intelligence (NI) is best understood as a feedback loop. Humans create AI, training it on data generated by human activity to enhance or replicate human capabilities. This AI
 AI's Biggest Secret — Creators Don't Understand It, Experts SplitMay 09, 2025 am 11:09 AM
AI's Biggest Secret — Creators Don't Understand It, Experts SplitMay 09, 2025 am 11:09 AMAnthropic's recent statement, highlighting the lack of understanding surrounding cutting-edge AI models, has sparked a heated debate among experts. Is this opacity a genuine technological crisis, or simply a temporary hurdle on the path to more soph
 Bulbul-V2 by Sarvam AI: India's Best TTS ModelMay 09, 2025 am 10:52 AM
Bulbul-V2 by Sarvam AI: India's Best TTS ModelMay 09, 2025 am 10:52 AMIndia is a diverse country with a rich tapestry of languages, making seamless communication across regions a persistent challenge. However, Sarvam’s Bulbul-V2 is helping to bridge this gap with its advanced text-to-speech (TTS) t


Hot AI Tools

Undresser.AI Undress
AI-powered app for creating realistic nude photos

AI Clothes Remover
Online AI tool for removing clothes from photos.

Undress AI Tool
Undress images for free

Clothoff.io
AI clothes remover

Video Face Swap
Swap faces in any video effortlessly with our completely free AI face swap tool!

Hot Article

Hot Tools

Safe Exam Browser
Safe Exam Browser is a secure browser environment for taking online exams securely. This software turns any computer into a secure workstation. It controls access to any utility and prevents students from using unauthorized resources.

SublimeText3 Mac version
God-level code editing software (SublimeText3)

mPDF
mPDF is a PHP library that can generate PDF files from UTF-8 encoded HTML. The original author, Ian Back, wrote mPDF to output PDF files "on the fly" from his website and handle different languages. It is slower than original scripts like HTML2FPDF and produces larger files when using Unicode fonts, but supports CSS styles etc. and has a lot of enhancements. Supports almost all languages, including RTL (Arabic and Hebrew) and CJK (Chinese, Japanese and Korean). Supports nested block-level elements (such as P, DIV),

Notepad++7.3.1
Easy-to-use and free code editor

WebStorm Mac version
Useful JavaScript development tools






