Home >Common Problem >What is bootstrap test
What is bootstrap test
- 藏色散人Original
- 2019-07-31 14:02:2921057browse

What is the bootstrap test?
The bootstrap test is the bootstrap method test, which is the replacement sampling statistical method , by repeatedly sampling the data set multiple times, multiple evolutionary trees are constructed to check the branch credibility of a given tree.
Introduction to bootstrapping
The gene tree is an estimate of the true pattern of the evolutionary relationship of a set of sequences. It is said that the gene tree is an estimate because There is random variation in the number of substitutions, so the true gene tree is unknown. It can be expected that in the gene tree, short branches are less reliable than long branches.
So what criteria can be used to evaluate a certain gene tree? What about the reliability of a particular branching order? For example, in Figure 17.1C, can those data really separate the Mo/Ha lineage from the common ancestor that preceded the Hu/Ba/Co/Sh lineage?
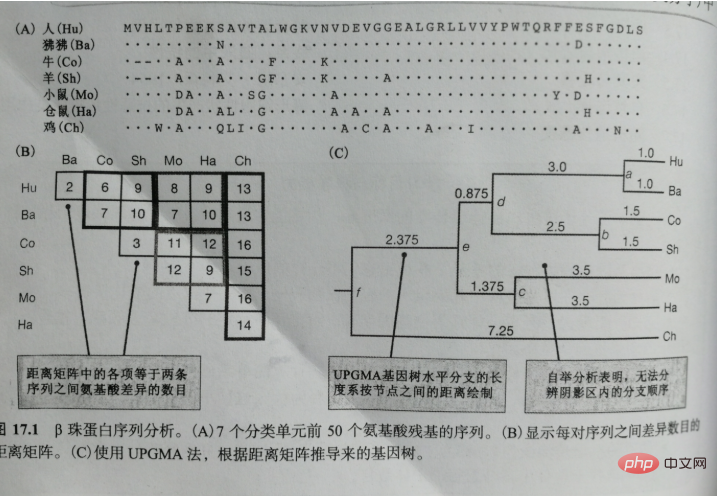
A common method to evaluate the reliability of a node in a gene tree is called bootstrapping. In this method, sites are randomly selected and constructed from actual data. Out of 1000 or more different data sets. Bootstrap sampling is performed with replacement sampling, which means that the same locus can be selected twice or more by chance. Therefore , a bootstrap sample from the sequence in Figure 17.1A is a sample of 50 sites randomly selected with replacement. In a specific bootstrap sample with a capacity of 50, 18 sites are expected to be Appear once, 9 sites will appear 2 times, 5 sites will appear 3 or more times, and 18 sites will not appear at all. Therefore, if in the gene tree A certain branching pattern is supported by most sites in the sequence, then the gene tree obtained from most bootstrap samples will contain the same branching pattern, but if the number of sites supporting a certain branching pattern is relatively small, then the gene tree derived from The gene trees of many bootstrap samples will not include this branching pattern.
In the gene tree in Figure 17.1C, out of 1000 bootstrap samples, fewer than 50 samples support the branching order included in the shaded area. %. Practically speaking, this result indicates that, for this small segment of the protein, the Hu/Ba, Co/Sh, and Mo/Ha taxa separated at very close times, and the question of which taxon separated first cannot be resolved. .
The above is the detailed content of What is bootstrap test. For more information, please follow other related articles on the PHP Chinese website!

