 Technology peripherals
Technology peripherals AI
AI Designing antibodies from scratch, Tencent and Peking University teams pre-trained large language models and published them in Nature sub-journal
Designing antibodies from scratch, Tencent and Peking University teams pre-trained large language models and published them in Nature sub-journal 编辑 | KX
编辑 | KX
AI 技术在辅助抗体设计方面取得了巨大进步。然而,抗体设计仍然严重依赖于从血清中分离抗原特异性抗体,这是一个资源密集且耗时的过程。
为了解决这个问题,腾讯 AI Lab、北京大学深圳研究生院和西京消化病医院研究团队提出了一种预训练抗体生成大语言模型(PALM-H3),用于从头生成具有所需抗原结合特异性的人工抗体CDRH3,减少对天然抗体的依赖。
此外,还设计了一个高精度的抗原-抗体结合预测模型 A2binder,将抗原表位序列与抗体序列配对,从而预测结合特异性和亲和力。
总之,该研究建立了一个用于抗体生成和评估的人工智能框架,这有可能显着加速抗体药物的开发。
相关研究以「De novo generation of SARS-CoV-2 antibody CDRH3 with a pre-trained generative large language model」为题,于 8 月 10 日发布在《Nature Communications》上。
抗体药物,又称单克隆抗体,在生物治疗中发挥着至关重要的作用。通过模仿免疫系统的作用,这些药物可以选择性地针对病毒和癌细胞等致病因子。与传统治疗方法相比,抗体药物是一种更具体、更有效的方法。抗体药物在治疗多种疾病方面已显示出积极的效果。
开发抗体药物是一个复杂的过程,包括从动物源中分离抗体,使其人性化,并优化其亲和力。但抗体药物的开发仍然严重依赖于天然抗体。
蛋白质的序列数据可以看作是一种语言,因此自然语言处理(NLP)领域的大规模预训练模型已被用来学习蛋白质的表征模式。当前已经开发了多种蛋白质语言模型。然而,由于抗体的多样性高和可用的抗原抗体配对数据稀缺,生成对特定抗原表位具有高亲和力的抗体仍然是一项具有挑战性的任务。
为了应对上述挑战,腾讯AI Lab 团队提出了预训练抗体生成大型语言模型PALM-H3,用于优化和生成重链互补决定区3 (CDRH3),该区域在抗体的特异性和多样性中起着至关重要的作用。
为了评估 PALM-H3 产生的抗体对抗原的亲和力,研究人员结合使用了抗原抗体对接和基于 AI 的方法。
研究人员还开发了用于评估抗体-抗原亲和力的 A2binder。 A2binder 能够实现准确且可推广的亲和力预测,即使对于未知抗原也是如此。
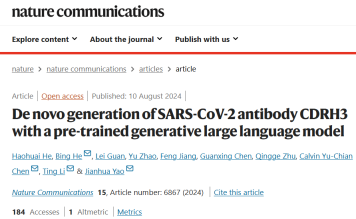
PALM-H3 和 A2Binder 的框架
PALM-H3 和 A2binder 的工作流程和模型框架如下图所示。
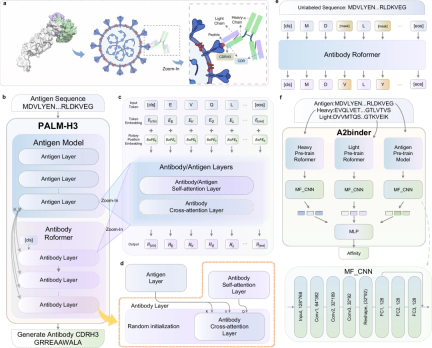
PALM-H3 的目的是生成抗体中的从头 CDRH3 序列。 CDRH3 区域在决定抗体对特定抗原序列的结合特异性方面起着最重要的作用。 PALM-H3 是一个类似 transformer 的模型,它使用基于 ESM2 的抗原模型作为编码器,使用抗体 Roformer 作为解码器。研究还构建了 A2binder 来预测人工生成的抗体的结合亲和力。
PALM-H3 和 A2binder 的构建包括三个步骤:首先,研究人员分别在未配对的抗体重链和轻链序列上预训练两个 Roformer 模型。然后,基于预训练的 ESM2、抗体重链 Roformer 和抗体轻链 Roformer 构建 A2binder,并使用配对亲和力数据对其进行训练。最后,使用预训练的 ESM2 和抗体重链 Roformer 构建 PALM-H3,并在配对抗原-CDRH3 数据上对其进行训练,以从头生成 CDRH3。
A2binder 可以准确预测抗原抗体结合概率、亲和力
通过将 A2binder 预测亲和力的能力与几种基线方法进行比较来评估其性能。
A2binder 在亲和力数据集上表现出色,部分原因在于抗体序列的预训练,这使得 A2binder 能够学习这些序列中存在的独特模式。
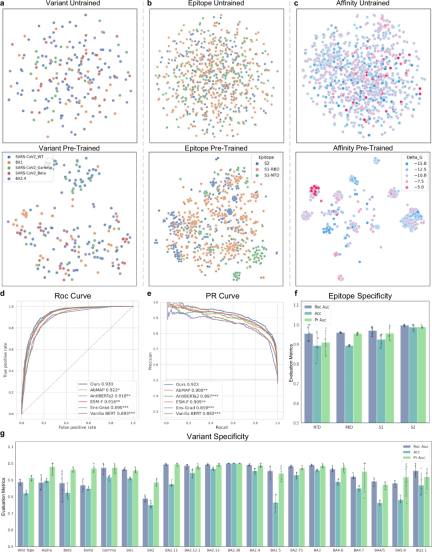
The results show that A2binder performs better than the baseline model ESM-F (the latter has the same framework, but the pre-trained model is replaced by ESM2) on all antigen-antibody affinity prediction datasets, which shows Pretraining with antibody sequences may be beneficial for related downstream tasks.
To evaluate the model’s performance in predicting affinity values, the researchers also utilized two datasets, 14H and 14L, that contain affinity value labels.
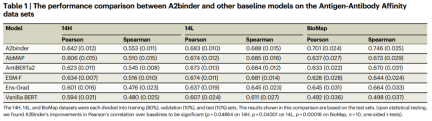
A2binder outperforms all baseline models on both Pearson correlation and Spearman correlation metrics. A2binder achieves a Pearson correlation of 0.642 on the 14H dataset (an improvement of 3%) and 0.683 on the 14L dataset (an improvement of 1%).
However, the performance of A2binder and other baseline models dropped slightly on the 14H and 14L datasets compared to other datasets. This observation is consistent with previous studies.
PALM-H3 excels in generating high-binding probability antibodies
Researchers explored the differences between the antibodies produced by PALM-H3 and natural antibodies. Their sequences were found to differ significantly, but the binding probabilities of the antibodies produced were not significantly affected by these differences. At the same time, their structural differences do lead to a decrease in binding affinity. These results are consistent with previous studies on network analysis of antibody libraries and generation of functional protein sequences.

Overall, the results show that PALM-H3 is capable of generating a diverse range of antibody sequences with high binding affinities, although unlike natural antibodies.
In addition, researchers verified the performance of PALM-H3 through ClusPro and SnugDock. PALM-H3 is capable of generating antibodies against a stabilizing peptide in the HR2 region of SARS-CoV-2, the CDRH3 sequence. It generated a novel CDRH3 sequence and validated that the generated sequence GRREAAWALA has improved targeting of antigen-stabilizing peptides compared to the native CDHR3 sequence GKAAGTFDS.
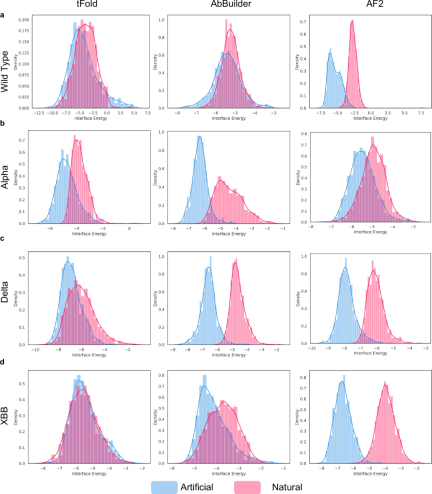
Additionally, PALM-H3 is able to generate antibodies with higher affinity against the emerging SARS-CoV-2 variant XBB CDRH3 sequence. The resulting sequence AKDSRTSPLRLDYS has a stronger affinity for XBB than its source, ASEVLDNLRDGYNF.
In addition, PALM-H3 not only overcomes the local optimal pitfalls faced by traditional sequential mutation strategies, but it also generates antibodies with higher antigen-binding affinity compared to the E-EVO approach. This highlights the advantages of PALM-H3 in antibody design, enabling more efficient exploration of sequence space and generation of high-affinity binders targeting specific epitopes.
In vitro experiments
In addition, the researchers also conducted in vitro experiments, including Western blotting, surface plasmon resonance analysis, and pseudovirus neutralization assays, providing key verification for the effectiveness of the PALM-H3 designed antibodies.
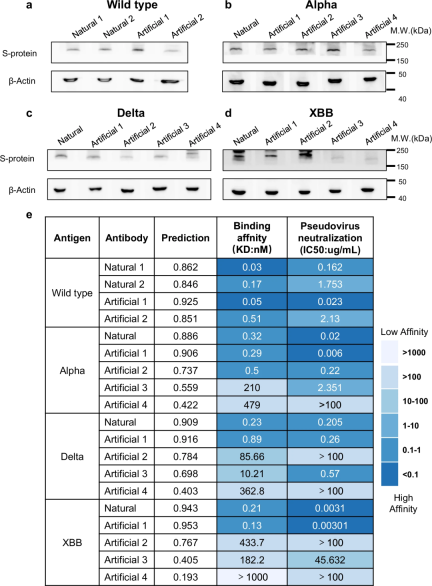
Two antibodies generated by PALM-H3 against SARS-CoV-2 wild-type, Alpha, Delta and XBB variant spike proteins achieved higher binding affinities than native antibodies in these experiments and neutralizing effect. The robust empirical results from these wet lab experiments complement computational predictions and analyses, validating the ability of PALM-H3 and A2binder to generate and select potent antibodies with high specificity and affinity for known and novel antigens.
In summary, the proposed PALM-H3 integrates the ability of large-scale antibody pre-training and the effectiveness of global feature fusion, resulting in excellent affinity prediction performance and the ability to design high-affinity antibodies. Furthermore, direct sequence generation and interpretable weight visualization make it an efficient and interpretable tool for designing high-affinity antibodies.
The above is the detailed content of Designing antibodies from scratch, Tencent and Peking University teams pre-trained large language models and published them in Nature sub-journal. For more information, please follow other related articles on the PHP Chinese website!
 五个时间序列预测的深度学习模型对比总结May 05, 2023 pm 05:16 PM
五个时间序列预测的深度学习模型对比总结May 05, 2023 pm 05:16 PMMakridakisM-Competitions系列(分别称为M4和M5)分别在2018年和2020年举办(M6也在今年举办了)。对于那些不了解的人来说,m系列得比赛可以被认为是时间序列生态系统的一种现有状态的总结,为当前得预测的理论和实践提供了经验和客观的证据。2018年M4的结果表明,纯粹的“ML”方法在很大程度上胜过传统的统计方法,这在当时是出乎意料的。在两年后的M5[1]中,最的高分是仅具有“ML”方法。并且所有前50名基本上都是基于ML的(大部分是树型模型)。这场比赛看到了LightG
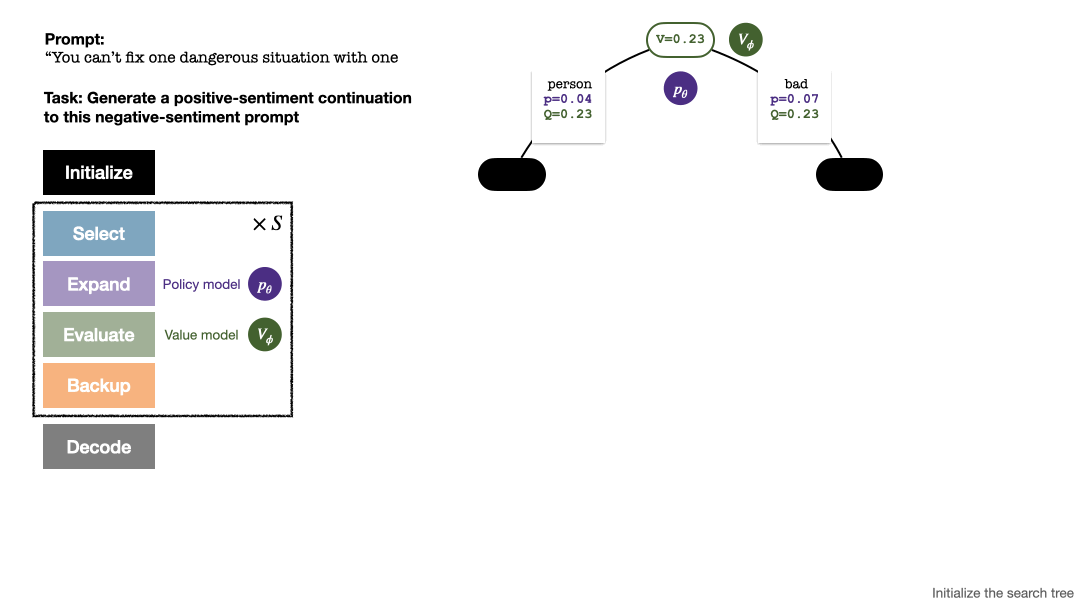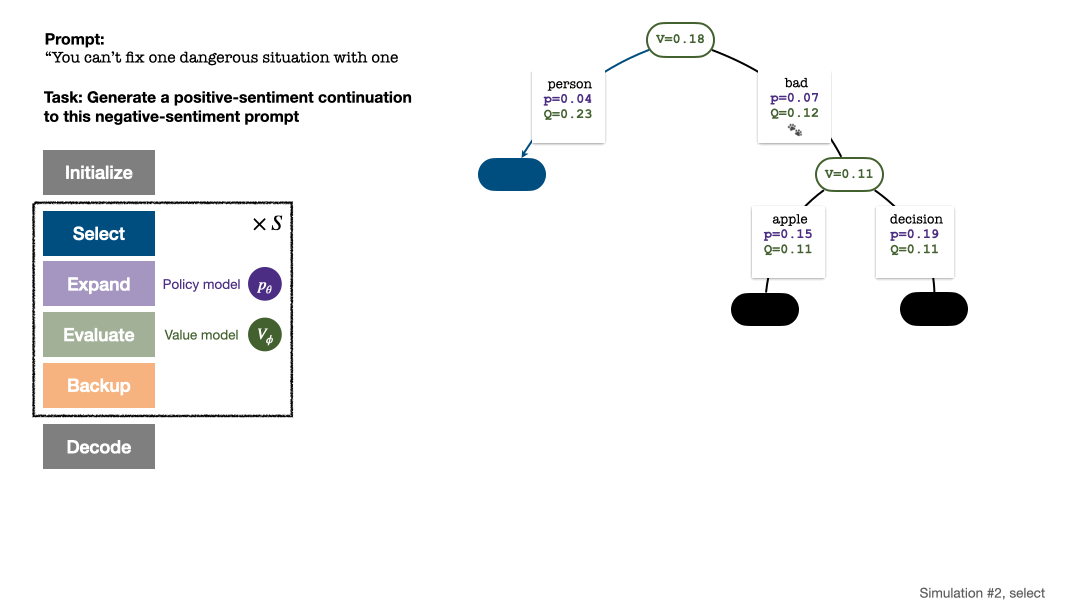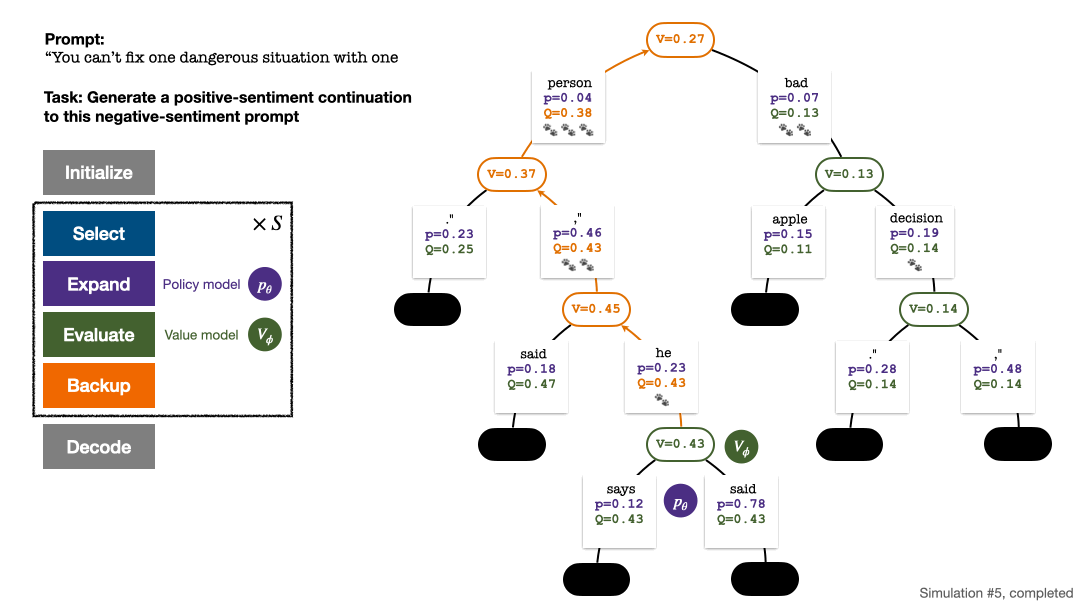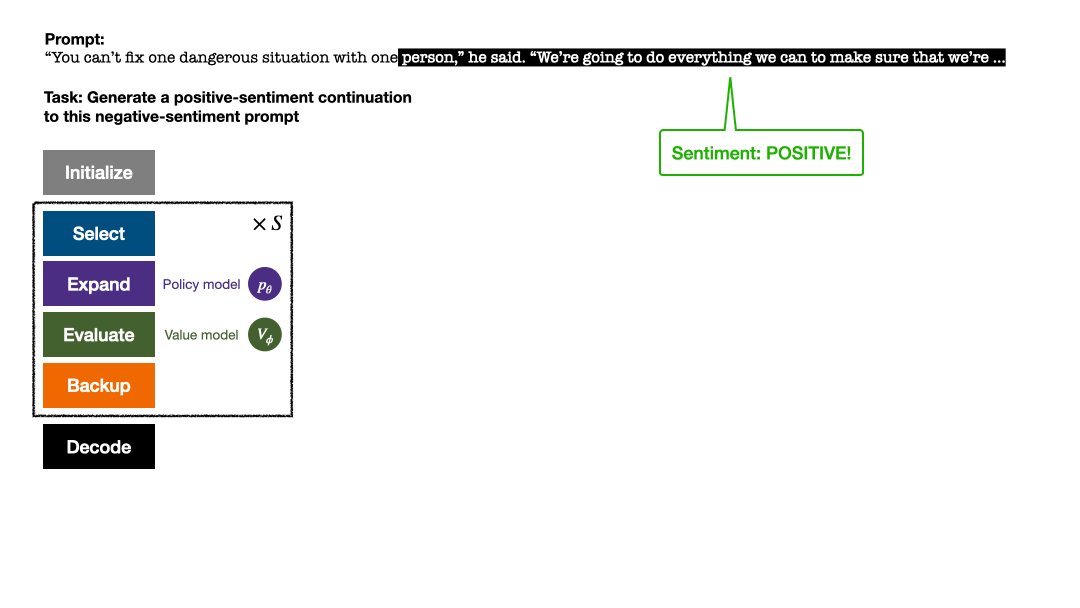 RLHF与AlphaGo核心技术强强联合,UW/Meta让文本生成能力再上新台阶Oct 27, 2023 pm 03:13 PM
RLHF与AlphaGo核心技术强强联合,UW/Meta让文本生成能力再上新台阶Oct 27, 2023 pm 03:13 PM在一项最新的研究中,来自UW和Meta的研究者提出了一种新的解码算法,将AlphaGo采用的蒙特卡洛树搜索算法(Monte-CarloTreeSearch,MCTS)应用到经过近端策略优化(ProximalPolicyOptimization,PPO)训练的RLHF语言模型上,大幅提高了模型生成文本的质量。PPO-MCTS算法通过探索与评估若干条候选序列,搜索到更优的解码策略。通过PPO-MCTS生成的文本能更好满足任务要求。论文链接:https://arxiv.org/pdf/2309.150
 MIT团队运用机器学习闭环自主分子发现平台,成功发现、合成和描述了303种新分子Jan 04, 2024 pm 05:38 PM
MIT团队运用机器学习闭环自主分子发现平台,成功发现、合成和描述了303种新分子Jan 04, 2024 pm 05:38 PM编辑|X传统意义上,发现所需特性的分子过程一直是由手动实验、化学家的直觉以及对机制和第一原理的理解推动的。随着化学家越来越多地使用自动化设备和预测合成算法,自主研究设备越来越接近实现。近日,来自MIT的研究人员开发了由集成机器学习工具驱动的闭环自主分子发现平台,以加速具有所需特性的分子的设计。无需手动实验即可探索化学空间并利用已知的化学结构。在两个案例研究中,该平台尝试了3000多个反应,其中1000多个产生了预测的反应产物,提出、合成并表征了303种未报道的染料样分子。该研究以《Autonom
 AI助力脑机接口研究,纽约大学突破性神经语音解码技术,登Nature子刊Apr 17, 2024 am 08:40 AM
AI助力脑机接口研究,纽约大学突破性神经语音解码技术,登Nature子刊Apr 17, 2024 am 08:40 AM作者|陈旭鹏编辑|ScienceAI由于神经系统的缺陷导致的失语会导致严重的生活障碍,它可能会限制人们的职业和社交生活。近年来,深度学习和脑机接口(BCI)技术的飞速发展为开发能够帮助失语者沟通的神经语音假肢提供了可行性。然而,神经信号的语音解码面临挑战。近日,约旦大学VideoLab和FlinkerLab的研究者开发了一个新型的可微分语音合成器,可以利用一个轻型的卷积神经网络将语音编码为一系列可解释的语音参数(例如音高、响度、共振峰频率等),并通过可微分神经网络将这些参数合成为语音。这个合成器
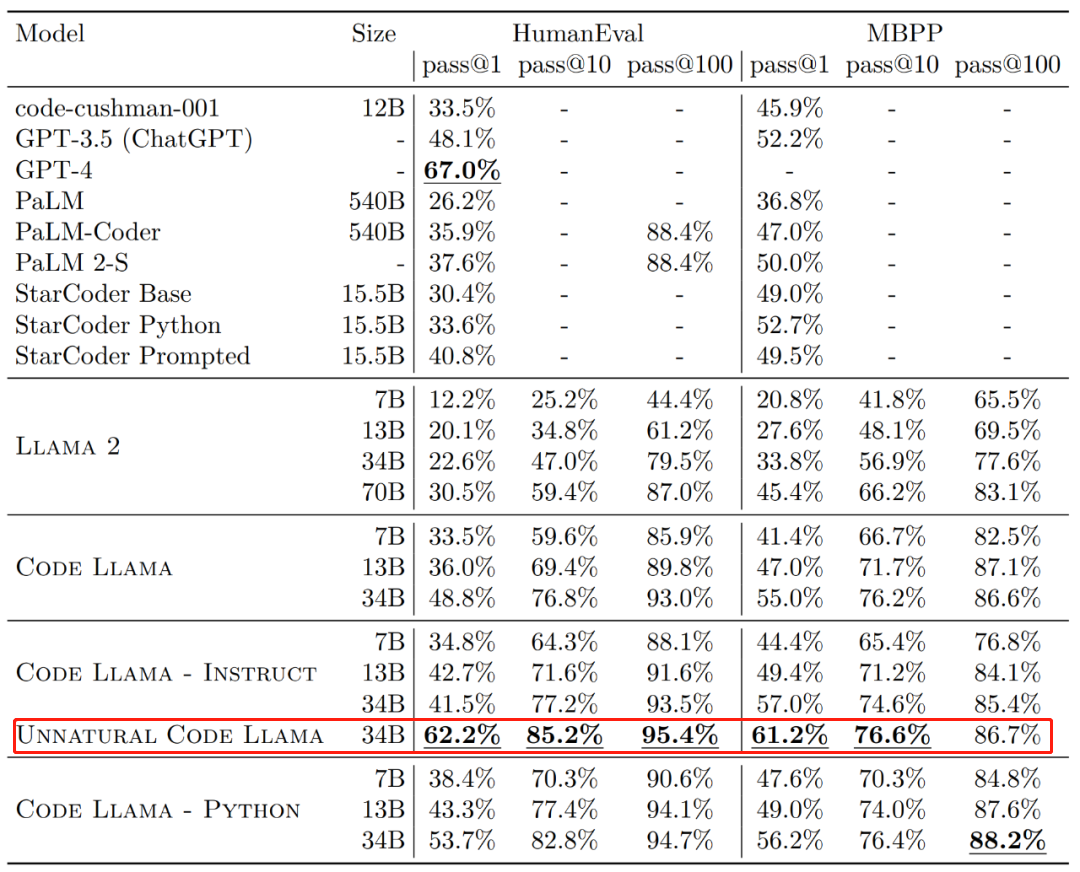 Code Llama代码能力飙升,微调版HumanEval得分超越GPT-4,一天发布Aug 26, 2023 pm 09:01 PM
Code Llama代码能力飙升,微调版HumanEval得分超越GPT-4,一天发布Aug 26, 2023 pm 09:01 PM昨天,Meta开源专攻代码生成的基础模型CodeLlama,可免费用于研究以及商用目的。CodeLlama系列模型有三个参数版本,参数量分别为7B、13B和34B。并且支持多种编程语言,包括Python、C++、Java、PHP、Typescript(Javascript)、C#和Bash。Meta提供的CodeLlama版本包括:代码Llama,基础代码模型;代码羊-Python,Python微调版本;代码Llama-Instruct,自然语言指令微调版就其效果来说,CodeLlama的不同版
 手机摄影技术让以假乱真的好莱坞级电影特效视频走红Sep 07, 2023 am 09:41 AM
手机摄影技术让以假乱真的好莱坞级电影特效视频走红Sep 07, 2023 am 09:41 AM一个普通人用一台手机就能制作电影特效的时代已经来了。最近,一个名叫Simulon的3D技术公司发布了一系列特效视频,视频中的3D机器人与环境无缝融合,而且光影效果非常自然。呈现这些效果的APP也叫Simulon,它能让使用者通过手机摄像头的实时拍摄,直接渲染出CGI(计算机生成图像)特效,就跟打开美颜相机拍摄一样。在具体操作中,你要先上传一个3D模型(比如图中的机器人)。Simulon会将这个模型放置到你拍摄的现实世界中,并使用准确的照明、阴影和反射效果来渲染它们。整个过程不需要相机解算、HDR
 准确率 >98%,基于电子密度的 GPT 用于化学研究,登 Nature 子刊Mar 27, 2024 pm 02:16 PM
准确率 >98%,基于电子密度的 GPT 用于化学研究,登 Nature 子刊Mar 27, 2024 pm 02:16 PM编辑|紫罗可合成分子的化学空间是非常广阔的。有效地探索这个领域需要依赖计算筛选技术,比如深度学习,以便快速地发现各种有趣的化合物。将分子结构转换为数字表示形式,并开发相应算法生成新的分子结构是进行化学发现的关键。最近,英国格拉斯哥大学的研究团队提出了一种基于电子密度训练的机器学习模型,用于生成主客体binders。这种模型能够以简化分子线性输入规范(SMILES)格式读取数据,准确率高达98%,从而实现对分子在二维空间的全面描述。通过变分自编码器生成主客体系统的电子密度和静电势的三维表示,然后通
 NVIDIA、Mila、Caltech联合发布LLM结合药物发现的多模态分子结构-文本模型Jan 14, 2024 pm 08:00 PM
NVIDIA、Mila、Caltech联合发布LLM结合药物发现的多模态分子结构-文本模型Jan 14, 2024 pm 08:00 PM作者|刘圣超编辑|凯霞从2021年开始,大语言和多模态的结合席卷了机器学习科研界。随着大模型和多模态应用的发展,我们是否可以将这些技术应用于药物发现呢?而且,这些自然语言的文本描述是否可以为这个具有挑战性的问题带来新的视角呢?答案是肯定的,并且我们对此持乐观态度近日,加拿大蒙特利尔学习算法研究院(Mila)、NVIDIAResearch、伊利诺伊大学厄巴纳-香槟分校(UIUC)、普林斯顿大学和加州理工学院的研究团队,通过对比学习策略共同学习分子的化学结构和文本描述,提出了一种多模态分子结构-文本


Hot AI Tools

Undresser.AI Undress
AI-powered app for creating realistic nude photos

AI Clothes Remover
Online AI tool for removing clothes from photos.

Undress AI Tool
Undress images for free

Clothoff.io
AI clothes remover

AI Hentai Generator
Generate AI Hentai for free.

Hot Article

Hot Tools

Atom editor mac version download
The most popular open source editor

VSCode Windows 64-bit Download
A free and powerful IDE editor launched by Microsoft

MinGW - Minimalist GNU for Windows
This project is in the process of being migrated to osdn.net/projects/mingw, you can continue to follow us there. MinGW: A native Windows port of the GNU Compiler Collection (GCC), freely distributable import libraries and header files for building native Windows applications; includes extensions to the MSVC runtime to support C99 functionality. All MinGW software can run on 64-bit Windows platforms.

SublimeText3 Linux new version
SublimeText3 Linux latest version

mPDF
mPDF is a PHP library that can generate PDF files from UTF-8 encoded HTML. The original author, Ian Back, wrote mPDF to output PDF files "on the fly" from his website and handle different languages. It is slower than original scripts like HTML2FPDF and produces larger files when using Unicode fonts, but supports CSS styles etc. and has a lot of enhancements. Supports almost all languages, including RTL (Arabic and Hebrew) and CJK (Chinese, Japanese and Korean). Supports nested block-level elements (such as P, DIV),






