 Technology peripherals
Technology peripherals AI
AI SOTA performance, University of Washington developed Transformer model to convert mass spectra into peptide sequences, published in Nature sub-journal
SOTA performance, University of Washington developed Transformer model to convert mass spectra into peptide sequences, published in Nature sub-journalSOTA performance, University of Washington developed Transformer model to convert mass spectra into peptide sequences, published in Nature sub-journal

A fundamental challenge in mass spectrometry-based proteomics is the identification of the peptides generating each tandem mass spectrum (MS/MS). Methods that rely on databases of known peptide sequences are unable to detect unexpected peptides and may be impractical or unapplicable in some cases.
Thus, the ability to assign peptide sequences into MS/MS without prior information (i.e. de novo peptide sequencing) is extremely valuable for tasks such as antibody sequencing, immunopeptidomics, and metaproteomics.
Although many methods have been developed to solve this problem, it remains an open challenge, partly due to the difficulty in modeling the irregular data structure of MS/MS.
Here, researchers at the University of Washington describe Casanovo, a machine learning model that uses the Transformer neural network architecture to convert peak sequences in MS/MS into the amino acid sequences that make up the resulting peptides.
The team trained the Casanovo model on 30 million labeled spectra and demonstrated that the model outperformed several state-of-the-art methods on cross-species benchmark datasets.
The team also developed a version of Casanovo fine-tuned for non-enzymatic peptides. This tool improves the analysis of immunopeptidomics and metaproteomics experiments and enables scientists to delve deeper into the dark proteome.
The study was titled "Sequence-to-sequence translation from mass spectra to peptides with a transformer model" and was published in "Nature Communications" on July 31, 2024.

- Tandem mass spectrometry (MS/MS) technology produces complex data, and converting these spectra into protein amino acid sequences is challenging.
- Deep learning has become the first choice for de novo peptide sequencing, but its limitations include: small number of annotated MS/MS spectra, difficulty in encoding high-resolution MS/MS data, complex neural networks and post-processing steps.
- Casanovo reframes the de novo peptide sequencing task as a machine translation problem, using the Transformer architecture to output predicted peptide sequences directly using m/z and intensity value pairs of MS/MS spectra.
-
In the latest research, Casanovo has made improvements, including:
- Expanded training set using 669 million spectra in the MassIVE-KB spectral library.
- Strict FDR control, searches data at 1% FDR, retaining only 100 PSMs for each unique precursor, for a total of 30 million high-quality PSMs.
- Beam search decoder that predicts the best peptide for each MS/MS spectrum.
## Casanovo: De novo peptide sequencing using the Transformer architecture
Figure 1: Casanovo performs de novo peptide sequencing using the Transformer architecture. (Source: Paper)
Casanovo’s outstanding performance is attributed to two aspects:
- Having a large amount of high-quality training data
- Using Transformer architecture
Transformer architecture
Transformer architecture is particularly suitable for converting variable lengths The elements of a sequence are placed in context and thus have great success in natural language modeling. Compared to recurrent neural networks, the Transformer architecture is capable of learning long-distance dependencies between sequence elements and can be parallelized for efficient training.
Applications of Casanovo
Casanovo encodes mass spectral peaks into sequences, taking advantage of the Transformer architecture and the rapid development of large language models to improve de novo peptide sequencing of MS/MS spectra.
Application scenarios:
- Paleoproteomics
- Forensic medicine
- Astrobiology
- Detection of peptides not present in the database
- As a post-processor for standard database searches
Antibody sequencing
Casanovo has not yet explored the use of antibody sequencing. However, a study by Denis Beslic's group at BAM in Germany conducted a systematic comparison of six de novo sequencing tools, including Casanovo, on the issue of antibody sequencing.

Novor, pNovo 3, DeepNovo, SMSNet, PointNovo and Casanovo for different enzymes on IgG1-Human-HC.
Related links:
https://academic.oup.com/bib/article/24/1/bbac542/6955273?login=false
Results:
Casanovo 在所有考慮指標上均明顯優於競爭方法。值得注意的是,此比較使用了貪婪解碼版本 Casanovo,並且僅對 200 萬個光譜進行訓練。
評估:
Casanovo 團隊對 Casanovo 進行了九種物種基準測試評估。下圖顯示,使用 3000 萬個光譜訓練的更新版本 Casanovo 可以產生更好的抗體定序性能。

未來,Casanovo 模型將有許多機會針對特定應用進行微調。研究人員對非酶模型的分析表明,Casanovo 的酶偏差可以透過使用相對較少的訓練數據進行調整。
因此,短期內,該團隊計劃訓練適用於各種不同裂解酶的 Casanovo 變體。 Casanovo 軟體使這種微調變得簡單,因此任何有興趣將模型調整到特定實驗設定的用戶都應該能夠這樣做。
從長遠來看,理想的模型將光譜以及相關元數據(例如消化酶、碰撞能量和儀器類型)作為輸入,並準確預測多種不同類型的實驗設置。
深度學習方法在提高從頭定序能力方面的潛力現已廣受認可。在論文接受審查期間,至少有六種其他深度學習從頭測序方法已發表,包括 GraphNovo、PepNet、Denovo-GCN、Spectralis、π-HelixNovo 和 NovoB。顯然,對這一不斷發展的工具領域進行全面而嚴格的基準比較將使該領域受益。
與此相關的是,現階段該領域的主要瓶頸之一是缺乏嚴格的從頭測序置信度評估方法。
在宏蛋白質組學分析中,研究人員將 Casanovo 預測與目標和相應的誘餌勝肽資料庫進行了匹配,但這種方法忽略了從頭測序將勝肽分配給外來譜的能力。
因此,一個懸而未決的問題是,對於給定的資料依賴型擷取資料集,Casanovo 是否在檢測勝肽的統計能力方面優於標準資料庫搜尋程序。
研究人員表示,透過足夠大的訓練集進行訓練,也許可以結束資料庫搜尋在 DDA 串聯質譜資料分析領域的統治地位。
論文連結:https://www.nature.com/articles/s41467-024-49731-x
The above is the detailed content of SOTA performance, University of Washington developed Transformer model to convert mass spectra into peptide sequences, published in Nature sub-journal. For more information, please follow other related articles on the PHP Chinese website!
 The AI Skills Gap Is Slowing Down Supply ChainsApr 26, 2025 am 11:13 AM
The AI Skills Gap Is Slowing Down Supply ChainsApr 26, 2025 am 11:13 AMThe term "AI-ready workforce" is frequently used, but what does it truly mean in the supply chain industry? According to Abe Eshkenazi, CEO of the Association for Supply Chain Management (ASCM), it signifies professionals capable of critic
 How One Company Is Quietly Working To Transform AI ForeverApr 26, 2025 am 11:12 AM
How One Company Is Quietly Working To Transform AI ForeverApr 26, 2025 am 11:12 AMThe decentralized AI revolution is quietly gaining momentum. This Friday in Austin, Texas, the Bittensor Endgame Summit marks a pivotal moment, transitioning decentralized AI (DeAI) from theory to practical application. Unlike the glitzy commercial
 Nvidia Releases NeMo Microservices To Streamline AI Agent DevelopmentApr 26, 2025 am 11:11 AM
Nvidia Releases NeMo Microservices To Streamline AI Agent DevelopmentApr 26, 2025 am 11:11 AMEnterprise AI faces data integration challenges The application of enterprise AI faces a major challenge: building systems that can maintain accuracy and practicality by continuously learning business data. NeMo microservices solve this problem by creating what Nvidia describes as "data flywheel", allowing AI systems to remain relevant through continuous exposure to enterprise information and user interaction. This newly launched toolkit contains five key microservices: NeMo Customizer handles fine-tuning of large language models with higher training throughput. NeMo Evaluator provides simplified evaluation of AI models for custom benchmarks. NeMo Guardrails implements security controls to maintain compliance and appropriateness
 AI Paints A New Picture For The Future Of Art And DesignApr 26, 2025 am 11:10 AM
AI Paints A New Picture For The Future Of Art And DesignApr 26, 2025 am 11:10 AMAI: The Future of Art and Design Artificial intelligence (AI) is changing the field of art and design in unprecedented ways, and its impact is no longer limited to amateurs, but more profoundly affecting professionals. Artwork and design schemes generated by AI are rapidly replacing traditional material images and designers in many transactional design activities such as advertising, social media image generation and web design. However, professional artists and designers also find the practical value of AI. They use AI as an auxiliary tool to explore new aesthetic possibilities, blend different styles, and create novel visual effects. AI helps artists and designers automate repetitive tasks, propose different design elements and provide creative input. AI supports style transfer, which is to apply a style of image
 How Zoom Is Revolutionizing Work With Agentic AI: From Meetings To MilestonesApr 26, 2025 am 11:09 AM
How Zoom Is Revolutionizing Work With Agentic AI: From Meetings To MilestonesApr 26, 2025 am 11:09 AMZoom, initially known for its video conferencing platform, is leading a workplace revolution with its innovative use of agentic AI. A recent conversation with Zoom's CTO, XD Huang, revealed the company's ambitious vision. Defining Agentic AI Huang d
 The Existential Threat To UniversitiesApr 26, 2025 am 11:08 AM
The Existential Threat To UniversitiesApr 26, 2025 am 11:08 AMWill AI revolutionize education? This question is prompting serious reflection among educators and stakeholders. The integration of AI into education presents both opportunities and challenges. As Matthew Lynch of The Tech Edvocate notes, universit
 The Prototype: American Scientists Are Looking For Jobs AbroadApr 26, 2025 am 11:07 AM
The Prototype: American Scientists Are Looking For Jobs AbroadApr 26, 2025 am 11:07 AMThe development of scientific research and technology in the United States may face challenges, perhaps due to budget cuts. According to Nature, the number of American scientists applying for overseas jobs increased by 32% from January to March 2025 compared with the same period in 2024. A previous poll showed that 75% of the researchers surveyed were considering searching for jobs in Europe and Canada. Hundreds of NIH and NSF grants have been terminated in the past few months, with NIH’s new grants down by about $2.3 billion this year, a drop of nearly one-third. The leaked budget proposal shows that the Trump administration is considering sharply cutting budgets for scientific institutions, with a possible reduction of up to 50%. The turmoil in the field of basic research has also affected one of the major advantages of the United States: attracting overseas talents. 35
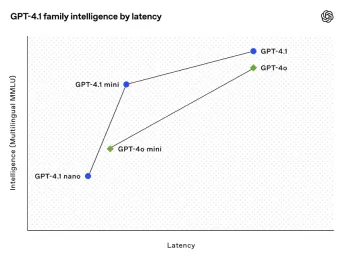 All About Open AI's Latest GPT 4.1 Family - Analytics VidhyaApr 26, 2025 am 10:19 AM
All About Open AI's Latest GPT 4.1 Family - Analytics VidhyaApr 26, 2025 am 10:19 AMOpenAI unveils the powerful GPT-4.1 series: a family of three advanced language models designed for real-world applications. This significant leap forward offers faster response times, enhanced comprehension, and drastically reduced costs compared t


Hot AI Tools

Undresser.AI Undress
AI-powered app for creating realistic nude photos

AI Clothes Remover
Online AI tool for removing clothes from photos.

Undress AI Tool
Undress images for free

Clothoff.io
AI clothes remover

Video Face Swap
Swap faces in any video effortlessly with our completely free AI face swap tool!

Hot Article

Hot Tools

Atom editor mac version download
The most popular open source editor

MantisBT
Mantis is an easy-to-deploy web-based defect tracking tool designed to aid in product defect tracking. It requires PHP, MySQL and a web server. Check out our demo and hosting services.

DVWA
Damn Vulnerable Web App (DVWA) is a PHP/MySQL web application that is very vulnerable. Its main goals are to be an aid for security professionals to test their skills and tools in a legal environment, to help web developers better understand the process of securing web applications, and to help teachers/students teach/learn in a classroom environment Web application security. The goal of DVWA is to practice some of the most common web vulnerabilities through a simple and straightforward interface, with varying degrees of difficulty. Please note that this software

WebStorm Mac version
Useful JavaScript development tools

SAP NetWeaver Server Adapter for Eclipse
Integrate Eclipse with SAP NetWeaver application server.






