Technology peripherals
Technology peripherals AI
AI Login to Science, drug affinity increased 37 times, AI performs unsupervised optimization of protein and antibody complexes
Login to Science, drug affinity increased 37 times, AI performs unsupervised optimization of protein and antibody complexes
Proteins are involved in many biological functions such as cell composition, muscle contraction, digestion of food, and identification of viruses.
In order to design better proteins (including antibodies), scientists often repeatedly mutate amino acids (arranging the units that make up proteins in a certain order) at different positions until the protein obtains the required function.
But there are more amino acid sequences than there are grains of sand in the world, so finding the best protein, and thus the best potential drug, is often difficult. When faced with this challenge, scientists often spend millions of dollars and test on miniaturized, simplified versions of biological systems.
“This requires a lot of guesswork and verification.” Brian L. Hie, assistant professor of chemical engineering at Stanford University and innovation fellow at the Arc Institute, said, “The goal of many intelligent algorithms is to take the guesswork out of it.”
Stanford University scientists have developed a new method based on machine learning that can predict molecular changes that lead to better antibody drugs faster and more accurately. Combining the 3D structure of the protein backbone with a large language model based on the amino acid sequence, the researchers were able to find rare and desirable mutations in minutes.
The study was titled "Unsupervised evolution of protein and antibody complexes with a structure-informed language model" and was published in "Science" on July 4, 2024.

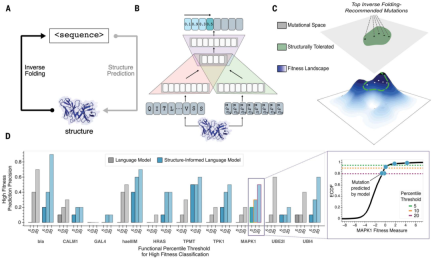
Large language models trained solely on sequence information can learn high-level principles of protein design. However, in addition to sequence, the three-dimensional structure of proteins determines their specific function, activity, and evolvability.
For antibody engineering problems, researchers at Stanford University applied structurally informed protein language models to predict high-fitness sequences constrained by known antibody or antibody-antigen complex structures.
Research shows that a universal protein language model augmented with protein structural backbone coordinates can guide the evolution of different proteins without the need to model individual functional tasks.
-
Structure-guided paradigm:
- does not model an explicit definition of protein function or fitness.
- Focus on regions that retain protein backbone folding and indirectly explore the fitness landscape.
- Assume evolution within a high sequence likelihood range is a valid prior for high fitness variants.
-
Wide application:
- can indirectly study the fitness landscape of proteins in different environments, such as enzyme catalysis, antibiotic resistance and chemotherapy resistance.
-
Protein complex design:
- ESM-IF1 trained only on single-chain structures can be extended to design protein complexes.
- Shows that structural information language models can implicitly learn to combine features and generalize to polyproteins.
-
Human Antibody Evolution:
- This method is particularly valuable for the evolution of human antibodies and can be used to treat a variety of diseases.
- Antibodies provide protection by binding to target antigens.
-
Replace large amounts of data:
- Structures can replace large amounts of data and the computer can still learn.
- More antibodies have optimization opportunities.
-
Directed evolution:
- This method is used to experimentally guide directed evolution activities of multiple proteins.
- Generate designs with functional activity superior to wild-type proteins.
- No need to analyze labeled fitness data or task-specific model supervision.
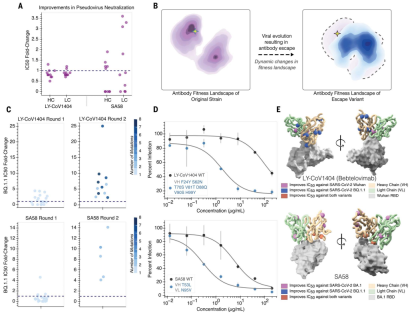
Illustration: Evolving antibodies using structural information language models can improve neutralizing potency and resilience. (Source: paper)
With this method, the team screened about 30 candidates for two therapeutic clinical antibodies for the treatment of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection. Variants. At the same time, the researchers achieved a 25-fold increase in neutralization and a 37-fold increase in affinity against BQ.1.1 and XBB.1.5 antibody-escape virus variants, respectively.
In conclusion, This tool will help quickly respond to new or developing diseases. It also lowers the barriers to making more effective drugs. Stronger drugs mean lower doses are needed, meaning more patients can benefit from a given dose.
論文連結:https://www.science.org/doi/10.1126/science.adk8946
相關報告:https://phys.org/news/2024-07-ai-approach-optimizes-approach-optimizes-approach-optimizes-approach-optimizes-approach-optimizes- antibody-drugs.html
The above is the detailed content of Login to Science, drug affinity increased 37 times, AI performs unsupervised optimization of protein and antibody complexes. For more information, please follow other related articles on the PHP Chinese website!
 2023年机器学习的十大概念和技术Apr 04, 2023 pm 12:30 PM
2023年机器学习的十大概念和技术Apr 04, 2023 pm 12:30 PM机器学习是一个不断发展的学科,一直在创造新的想法和技术。本文罗列了2023年机器学习的十大概念和技术。 本文罗列了2023年机器学习的十大概念和技术。2023年机器学习的十大概念和技术是一个教计算机从数据中学习的过程,无需明确的编程。机器学习是一个不断发展的学科,一直在创造新的想法和技术。为了保持领先,数据科学家应该关注其中一些网站,以跟上最新的发展。这将有助于了解机器学习中的技术如何在实践中使用,并为自己的业务或工作领域中的可能应用提供想法。2023年机器学习的十大概念和技术:1. 深度神经网
 人工智能自动获取知识和技能,实现自我完善的过程是什么Aug 24, 2022 am 11:57 AM
人工智能自动获取知识和技能,实现自我完善的过程是什么Aug 24, 2022 am 11:57 AM实现自我完善的过程是“机器学习”。机器学习是人工智能核心,是使计算机具有智能的根本途径;它使计算机能模拟人的学习行为,自动地通过学习来获取知识和技能,不断改善性能,实现自我完善。机器学习主要研究三方面问题:1、学习机理,人类获取知识、技能和抽象概念的天赋能力;2、学习方法,对生物学习机理进行简化的基础上,用计算的方法进行再现;3、学习系统,能够在一定程度上实现机器学习的系统。
 超参数优化比较之网格搜索、随机搜索和贝叶斯优化Apr 04, 2023 pm 12:05 PM
超参数优化比较之网格搜索、随机搜索和贝叶斯优化Apr 04, 2023 pm 12:05 PM本文将详细介绍用来提高机器学习效果的最常见的超参数优化方法。 译者 | 朱先忠审校 | 孙淑娟简介通常,在尝试改进机器学习模型时,人们首先想到的解决方案是添加更多的训练数据。额外的数据通常是有帮助(在某些情况下除外)的,但生成高质量的数据可能非常昂贵。通过使用现有数据获得最佳模型性能,超参数优化可以节省我们的时间和资源。顾名思义,超参数优化是为机器学习模型确定最佳超参数组合以满足优化函数(即,给定研究中的数据集,最大化模型的性能)的过程。换句话说,每个模型都会提供多个有关选项的调整“按钮
 得益于OpenAI技术,微软必应的搜索流量超过谷歌Mar 31, 2023 pm 10:38 PM
得益于OpenAI技术,微软必应的搜索流量超过谷歌Mar 31, 2023 pm 10:38 PM截至3月20日的数据显示,自微软2月7日推出其人工智能版本以来,必应搜索引擎的页面访问量增加了15.8%,而Alphabet旗下的谷歌搜索引擎则下降了近1%。 3月23日消息,外媒报道称,分析公司Similarweb的数据显示,在整合了OpenAI的技术后,微软旗下的必应在页面访问量方面实现了更多的增长。截至3月20日的数据显示,自微软2月7日推出其人工智能版本以来,必应搜索引擎的页面访问量增加了15.8%,而Alphabet旗下的谷歌搜索引擎则下降了近1%。这些数据是微软在与谷歌争夺生
 荣耀的人工智能助手叫什么名字Sep 06, 2022 pm 03:31 PM
荣耀的人工智能助手叫什么名字Sep 06, 2022 pm 03:31 PM荣耀的人工智能助手叫“YOYO”,也即悠悠;YOYO除了能够实现语音操控等基本功能之外,还拥有智慧视觉、智慧识屏、情景智能、智慧搜索等功能,可以在系统设置页面中的智慧助手里进行相关的设置。
 人工智能在教育领域的应用主要有哪些Dec 14, 2020 pm 05:08 PM
人工智能在教育领域的应用主要有哪些Dec 14, 2020 pm 05:08 PM人工智能在教育领域的应用主要有个性化学习、虚拟导师、教育机器人和场景式教育。人工智能在教育领域的应用目前还处于早期探索阶段,但是潜力却是巨大的。
 30行Python代码就可以调用ChatGPT API总结论文的主要内容Apr 04, 2023 pm 12:05 PM
30行Python代码就可以调用ChatGPT API总结论文的主要内容Apr 04, 2023 pm 12:05 PM阅读论文可以说是我们的日常工作之一,论文的数量太多,我们如何快速阅读归纳呢?自从ChatGPT出现以后,有很多阅读论文的服务可以使用。其实使用ChatGPT API非常简单,我们只用30行python代码就可以在本地搭建一个自己的应用。 阅读论文可以说是我们的日常工作之一,论文的数量太多,我们如何快速阅读归纳呢?自从ChatGPT出现以后,有很多阅读论文的服务可以使用。其实使用ChatGPT API非常简单,我们只用30行python代码就可以在本地搭建一个自己的应用。使用 Python 和 C
 人工智能在生活中的应用有哪些Jul 20, 2022 pm 04:47 PM
人工智能在生活中的应用有哪些Jul 20, 2022 pm 04:47 PM人工智能在生活中的应用有:1、虚拟个人助理,使用者可通过声控、文字输入的方式,来完成一些日常生活的小事;2、语音评测,利用云计算技术,将自动口语评测服务放在云端,并开放API接口供客户远程使用;3、无人汽车,主要依靠车内的以计算机系统为主的智能驾驶仪来实现无人驾驶的目标;4、天气预测,通过手机GPRS系统,定位到用户所处的位置,在利用算法,对覆盖全国的雷达图进行数据分析并预测。


Hot AI Tools

Undresser.AI Undress
AI-powered app for creating realistic nude photos

AI Clothes Remover
Online AI tool for removing clothes from photos.

Undress AI Tool
Undress images for free

Clothoff.io
AI clothes remover

AI Hentai Generator
Generate AI Hentai for free.

Hot Article

Hot Tools

WebStorm Mac version
Useful JavaScript development tools

SublimeText3 Linux new version
SublimeText3 Linux latest version

ZendStudio 13.5.1 Mac
Powerful PHP integrated development environment

SublimeText3 Mac version
God-level code editing software (SublimeText3)

SublimeText3 English version
Recommended: Win version, supports code prompts!