 Technology peripherals
Technology peripherals AI
AI In just a few seconds, protein dynamics information can be accurately inferred. AI models such as Shandong University and Beijing Institute of Technology RMSF-net are published in Nature sub-journals.
In just a few seconds, protein dynamics information can be accurately inferred. AI models such as Shandong University and Beijing Institute of Technology RMSF-net are published in Nature sub-journals.
The dynamics of a protein are crucial to understanding its mechanism. However, computationally predicting protein kinetic information is challenging.
Here, a research team from Shandong University, BioMap, Beijing Institute of Technology, Hubei Medical College, Ningxia Medical University and King Abdullah University of Science and Technology (KAUST) proposed a neural network model RMSF -net, which outperforms previous methods and produces the best results in large-scale protein dynamics data sets; the model can accurately infer the dynamics information of a protein in seconds.
By effectively learning from the integration of experimental protein structure data and cryo-EM data, this method is able to accurately identify interactive bidirectional constraints and supervision between cryo-EM images and PDB models to maximize Improve the efficiency of dynamics prediction.
RMSF-net is a free-to-use tool that will play an important role in protein dynamics studies.
The study was titled "Accurate Prediction of Protein Structural Flexibility by Deep Learning Integrating Intricate Atomic Structures and Cryo-EM Density Information" and was published in "Nature Communications" on July 2.

- https://www.nature.com/articles/s41467-024-49858-x
RMSF-net GitHub address:
- https://github. com/XintSong/RMSF-net
Protein Dynamics
Protein dynamics are crucial in understanding their mechanisms. Cryo-electron microscopy (cryo-EM) technology can resolve most proteins, where the macromolecular structure is represented by a 3D density map.
Limitations of cryo-electron microscopy
Due to the low resolution and signal-to-noise ratio of the original 2D particle images, cryo-electron microscopy analysis cannot resolve small conformational changes during reconstruction.
Application of deep learning in cryo-electron microscopy
Deep learning methods are widely used in the automatic analysis of cryo-electron microscopy images. Using high-resolution cryo-EM maps, a Protein Data Bank (PDB) model can be constructed from the cryo-EM maps.
RMSF-net Overview
RMSF-net is a neural network model for cryo-electron microscopy density maps. It leverages cryo-EM density and PDB model information to accurately infer protein dynamic information in seconds.
RMSF
RMSF is a widely used measurement method for assessing the flexibility of molecular structures in molecular dynamics (MD) analyses. Its main purpose is to predict the RMSF of local structures (residues, atoms) within a protein.
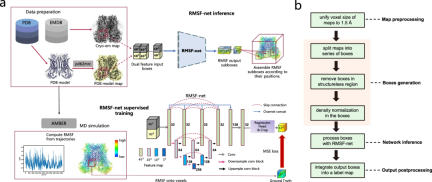
In addition to cryo-EM images, RMSF-net utilizes PDB models as additional input to produce RMSF predictions that are very close to the MD simulation results.
RMSF-net is a three-dimensional convolutional neural network containing two interconnected modules. The main module uses Unet+ (L3) architecture to encode and decode features of input density boxes. Another module utilizes 1x1 convolutions to regress the channels of the feature maps generated by the Unet+backbone. Center clipping is then applied to the regression module output to obtain a centered RMSF subbox, where the voxel value corresponds to the RMSF of the atoms contained within it. Finally, the RMSF subboxes are spatially merged into an RMSF map using a merging algorithm.
In addition, the researchers also constructed a large-scale protein dynamics dataset for training and validation of RMSF-net, in which 335 cryo-EM structural entries with fitted PDB models were selected and corresponding MD simulations were performed. Comprehensive experimental results demonstrate the efficiency and effectiveness of RMSF-net.
Table: Performance of different RMSF prediction methods on the data set. (Source: paper)

RMSF-net performed well in rigorous 5-fold cross-validation, with a correlation coefficient of 0.746±0.127 with MD simulation results. The correlation coefficient of RMSF-net is improved by 15% compared to DEFMap and by 10% compared to the baseline method.
Interpretability of dynamics predictions
Researchers enhanced the interpretability of RMSF-net dynamics predictions through comparative experiments. They divide the RMSF forecasting process into two steps:
- 結構資訊擷取(Occ2RMSF-net)
- 基於提取的結構資訊進行動力學預測
研究表明,基於低溫電子顯微鏡圖譜的模型(例如DEFMap 或RMSF-net_cryo)的動力學預測主要透過解讀蛋白質結構實現。這突顯了蛋白質拓樸結構與動力學之間的聯繫,符合結構-功能關係的第一原理。
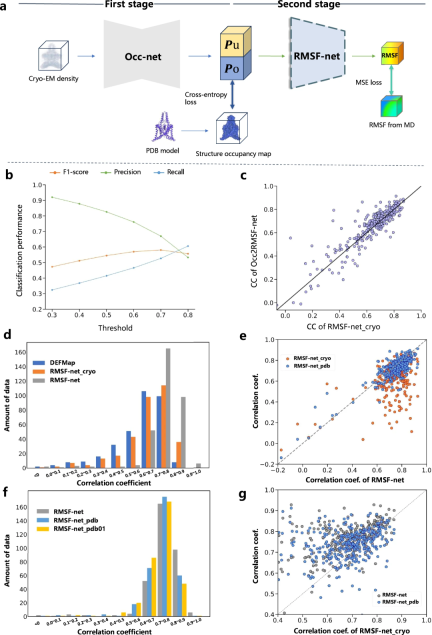
此外,透過對RMSF-net_cryo、RMSF-net_pdb 和最終的雙組合RMSF-net 進行全面比較,證明了:一方面,來自PDB 模型的結構資訊在RMSF-net 中起主要作用,其中深度模型從MD 模擬中學習結構拓撲和靈活性之間的模式,另一方面,低溫電子顯微鏡圖譜異質密度分佈中包含的動力學資訊進一步增強了模型。這些結果驗證了低溫電子顯微鏡圖和 PDB 模型的資訊對 RMSF-net 中的蛋白質動力學預測的互補作用。
局限性與未來方向
不可否認的是,RMSF-net 主要限於預測純蛋白質及其複合物在溶液中的柔韌性。對於蛋白質在與小分子配體結合或在膜環境中的動力學特性,該方法在某些局部區域可能會表現出不準確性。
RMSF-net 的卓越性能揭示了進一步研究該方向的可行性。該研究還沒有擴展到核酸和蛋白質-核酸複合物。綜合表徵大分子動力學的各個方面,包括多構象預測和轉變分析,在未來需要進一步進行廣泛而深入的研究。
儘管如此,作為預測蛋白質動力學的工具,RMSF-net 由於其優越的性能和超快的處理速度,在蛋白質結構和動力學研究中仍有很大的應用前景。
註:封面來自網路
The above is the detailed content of In just a few seconds, protein dynamics information can be accurately inferred. AI models such as Shandong University and Beijing Institute of Technology RMSF-net are published in Nature sub-journals.. For more information, please follow other related articles on the PHP Chinese website!
 Let's Dance: Structured Movement To Fine-Tune Our Human Neural NetsApr 27, 2025 am 11:09 AM
Let's Dance: Structured Movement To Fine-Tune Our Human Neural NetsApr 27, 2025 am 11:09 AMScientists have extensively studied human and simpler neural networks (like those in C. elegans) to understand their functionality. However, a crucial question arises: how do we adapt our own neural networks to work effectively alongside novel AI s
 New Google Leak Reveals Subscription Changes For Gemini AIApr 27, 2025 am 11:08 AM
New Google Leak Reveals Subscription Changes For Gemini AIApr 27, 2025 am 11:08 AMGoogle's Gemini Advanced: New Subscription Tiers on the Horizon Currently, accessing Gemini Advanced requires a $19.99/month Google One AI Premium plan. However, an Android Authority report hints at upcoming changes. Code within the latest Google P
 How Data Analytics Acceleration Is Solving AI's Hidden BottleneckApr 27, 2025 am 11:07 AM
How Data Analytics Acceleration Is Solving AI's Hidden BottleneckApr 27, 2025 am 11:07 AMDespite the hype surrounding advanced AI capabilities, a significant challenge lurks within enterprise AI deployments: data processing bottlenecks. While CEOs celebrate AI advancements, engineers grapple with slow query times, overloaded pipelines, a
 MarkItDown MCP Can Convert Any Document into Markdowns!Apr 27, 2025 am 09:47 AM
MarkItDown MCP Can Convert Any Document into Markdowns!Apr 27, 2025 am 09:47 AMHandling documents is no longer just about opening files in your AI projects, it’s about transforming chaos into clarity. Docs such as PDFs, PowerPoints, and Word flood our workflows in every shape and size. Retrieving structured
 How to Use Google ADK for Building Agents? - Analytics VidhyaApr 27, 2025 am 09:42 AM
How to Use Google ADK for Building Agents? - Analytics VidhyaApr 27, 2025 am 09:42 AMHarness the power of Google's Agent Development Kit (ADK) to create intelligent agents with real-world capabilities! This tutorial guides you through building conversational agents using ADK, supporting various language models like Gemini and GPT. W
 Use of SLM over LLM for Effective Problem Solving - Analytics VidhyaApr 27, 2025 am 09:27 AM
Use of SLM over LLM for Effective Problem Solving - Analytics VidhyaApr 27, 2025 am 09:27 AMsummary: Small Language Model (SLM) is designed for efficiency. They are better than the Large Language Model (LLM) in resource-deficient, real-time and privacy-sensitive environments. Best for focus-based tasks, especially where domain specificity, controllability, and interpretability are more important than general knowledge or creativity. SLMs are not a replacement for LLMs, but they are ideal when precision, speed and cost-effectiveness are critical. Technology helps us achieve more with fewer resources. It has always been a promoter, not a driver. From the steam engine era to the Internet bubble era, the power of technology lies in the extent to which it helps us solve problems. Artificial intelligence (AI) and more recently generative AI are no exception
 How to Use Google Gemini Models for Computer Vision Tasks? - Analytics VidhyaApr 27, 2025 am 09:26 AM
How to Use Google Gemini Models for Computer Vision Tasks? - Analytics VidhyaApr 27, 2025 am 09:26 AMHarness the Power of Google Gemini for Computer Vision: A Comprehensive Guide Google Gemini, a leading AI chatbot, extends its capabilities beyond conversation to encompass powerful computer vision functionalities. This guide details how to utilize
 Gemini 2.0 Flash vs o4-mini: Can Google Do Better Than OpenAI?Apr 27, 2025 am 09:20 AM
Gemini 2.0 Flash vs o4-mini: Can Google Do Better Than OpenAI?Apr 27, 2025 am 09:20 AMThe AI landscape of 2025 is electrifying with the arrival of Google's Gemini 2.0 Flash and OpenAI's o4-mini. These cutting-edge models, launched weeks apart, boast comparable advanced features and impressive benchmark scores. This in-depth compariso


Hot AI Tools

Undresser.AI Undress
AI-powered app for creating realistic nude photos

AI Clothes Remover
Online AI tool for removing clothes from photos.

Undress AI Tool
Undress images for free

Clothoff.io
AI clothes remover

Video Face Swap
Swap faces in any video effortlessly with our completely free AI face swap tool!

Hot Article

Hot Tools

ZendStudio 13.5.1 Mac
Powerful PHP integrated development environment

PhpStorm Mac version
The latest (2018.2.1) professional PHP integrated development tool

DVWA
Damn Vulnerable Web App (DVWA) is a PHP/MySQL web application that is very vulnerable. Its main goals are to be an aid for security professionals to test their skills and tools in a legal environment, to help web developers better understand the process of securing web applications, and to help teachers/students teach/learn in a classroom environment Web application security. The goal of DVWA is to practice some of the most common web vulnerabilities through a simple and straightforward interface, with varying degrees of difficulty. Please note that this software

WebStorm Mac version
Useful JavaScript development tools

SublimeText3 Chinese version
Chinese version, very easy to use





