 Technology peripherals
Technology peripherals AI
AI SOTA performance, multi-scale learning, Sun Yat-sen University proposes protein-drug interaction AI framework
SOTA performance, multi-scale learning, Sun Yat-sen University proposes protein-drug interaction AI frameworkSOTA performance, multi-scale learning, Sun Yat-sen University proposes protein-drug interaction AI framework

Editor | Ziluo
Interactions between proteins, drugs and other biomolecules play a vital role in various biological processes. Understanding these interactions is critical to deciphering the molecular mechanisms underlying biological processes and developing new therapeutic strategies. Proteins are among the most important molecules in cells, and they perform various functions within cells. Drugs often regulate physiological processes by interacting with specific proteins. These interactions can promote or inhibit specific molecular signaling pathways. Current multi-scale computational methods often rely too much on a single scale and under-fit other scales. This may be related to the uneven multi-scale tropism and inherent greed of multi-scale learning.
In order to alleviate the optimization imbalance, researchers from Sun Yat-sen University and Shanghai Jiao Tong University proposed a multi-scale representation learning framework MUSE based on variable expectation maximization, which can effectively integrate multi-scale information for learning. This strategy effectively fuses multi-scale information between the atomic structure and molecular network scales through mutual supervision and iterative optimization. This approach allows for greater information transfer and learning. This strategy effectively fuses multi-scale information between the atomic structure and molecular network scales through mutual supervision and iterative optimization.
MUSE+ not only outperforms the current state-of-the-art models on molecular interaction (protein-protein, drug-protein, and drug) tasks, but also outperforms the current state-of-the-art models on protein interface predictions at the atomic structure scale Model. More importantly, the multiscale learning framework can be extended to computational drug discovery at other scales.
The study, titled "
A variational expectation-maximization framework for balanced multi-scale learning of protein and drug interactions", was published in "Nature Communications" on May 25 "superior.
 Paper link:
Paper link:
Protein function is characterized by interactions with proteins, drugs, and other biomolecules. Understanding these interactions is critical for deciphering the molecular mechanisms of biological processes and developing new therapeutic strategies. However, the substantial growth in demands and costs associated with experimental interactions requires computational tools to automatically predict and understand interactions between biomolecules. To meet these demands and growing costs, computational tools are needed to automatically predict and understand interactions between biomolecules.
Predicting these interactions purely from structure is one of the most important challenges in structural biology. Current computational methods mostly predict interactions based on molecular networks or structural information and do not integrate them into a unified multiscale framework.
While some multi-view learning methods strive to fuse multi-scale information, an intuitive way to learn multi-scale representations is to combine molecular graphs with interaction networks and jointly optimize them. However, due to the imbalance and inherent greediness of multi-scale learning, these models often rely heavily on a single scale. Inability to effectively utilize information related to all scales and generalization is poor.
In addition, an effective multi-scale framework not only needs to capture rich information within different scales, but also needs to well preserve the potential relationships between them.
MUSE is used to learn multi-scale information of proteins and drugsHere, the Sun Yat-sen University research team proposed MUSE, a method based on variable expectation maximization (Expectation Maximization) A multi-scale representation learning framework that can optimize different scales in an alternating process of multiple iterations. Compared with existing methods that rely heavily on single-scale information, MUSE effectively solves the optimization imbalance problem in multi-scale learning through mutual supervision and iterative optimization.
MUSE is a multi-scale learning method that combines molecular structure modeling and protein-drug interaction network learning through a variational expectation maximization (EM) framework. The EM framework optimizes two modules, namely the expectation step (E-step) and the maximization step (M-step), in an alternating process of multiple iterations.
During the E-step, MUSE utilizes the structural information of each biomolecule to learn a valid structural representation for training in the M-step using known interactions and reinforcement samples. It takes as input protein and drug pairs and their atomic-level structural information, augmented by M-step predicted interactions. M-step takes as input the predicted interactions of the molecular-level interaction network, structural embeddings, and E-step and outputs the predicted interactions. Iterative optimization between E-step and M-step ensures interactive capture of molecular structure and network information with different learning rates at the two scales.
Mutual supervision ensures that each scale model learns in an appropriate manner, enabling it to exploit effective information at different scales. This framework will be demonstrated across multiple multiscales of interactions between proteins and drugs. It is analyzed that MUSE alleviates the imbalanced features in multi-scale learning and effectively integrates hierarchical and complementary information from different scales.

Outperforms current state-of-the-art models on multi-scale tasks
Utilizes atomic structure information to improve predictions at the molecular network scale
To evaluate their approach, the researchers first used MUSE to integrate atomic structure information to improve molecular network scale predictions. MUSE achieves state-of-the-art performance on three multi-scale interaction prediction tasks: protein-protein interaction (PPI), drug-protein interaction (DPI), and drug-drug interaction (DDI).

Improving atomic structure-scale predictions from the molecular network scale
In addition to using atomic structure information to improve molecular network-scale predictions, the researchers went one step further The ability of MUSE to learn and predict structural properties at the atomic structural scale was investigated, including predicting interfacial contacts and binding sites relevant to PPIs.
To evaluate predictions of protein interchain contacts, MUSE was compared to state-of-the-art methods on the DIPS-Plus benchmark. MUSE consistently outperforms all other methods, validating its effectiveness and adaptability in atomic structure prediction.

MUSE was further evaluated to predict whether residues are directly involved in protein-protein interactions. The results demonstrate that molecular network-scale learning in MUSE can provide valuable insights into atomic structure-scale predictions.
Alleviating the imbalance characteristics of multi-scale learning through iterative optimization
In order to explore why MUSE can achieve superior performance in multi-scale representation, researchers focused on the imbalance of multi-scale learning. The balancing characteristic analyzes MUSE's learning ability.

The results show that MUSE effectively alleviates the imbalance characteristics and greedy learning in multi-scale learning, ensuring the comprehensive utilization of information at different scales during the training process. In addition, experiments with utilization rate analysis allowed researchers to understand specifically what the model learned and demonstrated that using MUSE to balance model learning at different scales can enhance generalization capabilities.
Visualization and interpretation of learned multi-scale representations
To better understand the learned multi-scale representations, researchers have studied MUSE learning from different perspectives The multi-scale representation obtained includes (1) MUSE's ability to capture the atomic structure information (i.e., structural motifs and embeddings) involved in PPI, and (2) mutual supervision between the learned atomic structure and molecular network representations.

As an example of binding site prediction (PDB id: 3CQQ-A), MUSE can accurately identify residues belonging to the binding site with 97.7% accuracy. This suggests that mutual supervision in MUSE helps atomic structural scale models learn key substructures relevant to interactions.
Finally, the researchers also conducted ablation studies to study the impact of pseudo-labels predicted at the atomic structure scale on the molecular network scale.
While MUSE demonstrates state-of-the-art performance in benchmarks, it is still possible to improve its ability to handle noisy and incomplete multi-scale downstream tasks. This can be achieved by combining prior knowledge through knowledge graphs and explainable AI techniques. On the other hand, this conceptual multiscale framework can also be extended to computational drug discovery at other scales.
The above is the detailed content of SOTA performance, multi-scale learning, Sun Yat-sen University proposes protein-drug interaction AI framework. For more information, please follow other related articles on the PHP Chinese website!
 Personal Hacking Will Be A Pretty Fierce BearMay 11, 2025 am 11:09 AM
Personal Hacking Will Be A Pretty Fierce BearMay 11, 2025 am 11:09 AMCyberattacks are evolving. Gone are the days of generic phishing emails. The future of cybercrime is hyper-personalized, leveraging readily available online data and AI to craft highly targeted attacks. Imagine a scammer who knows your job, your f
 Pope Leo XIV Reveals How AI Influenced His Name ChoiceMay 11, 2025 am 11:07 AM
Pope Leo XIV Reveals How AI Influenced His Name ChoiceMay 11, 2025 am 11:07 AMIn his inaugural address to the College of Cardinals, Chicago-born Robert Francis Prevost, the newly elected Pope Leo XIV, discussed the influence of his namesake, Pope Leo XIII, whose papacy (1878-1903) coincided with the dawn of the automobile and
 FastAPI-MCP Tutorial for Beginners and Experts - Analytics VidhyaMay 11, 2025 am 10:56 AM
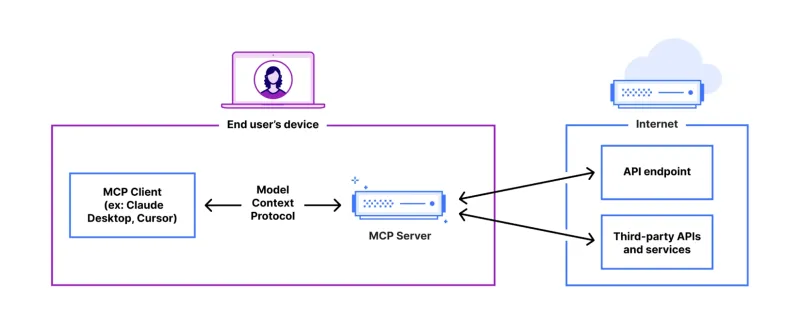
FastAPI-MCP Tutorial for Beginners and Experts - Analytics VidhyaMay 11, 2025 am 10:56 AMThis tutorial demonstrates how to integrate your Large Language Model (LLM) with external tools using the Model Context Protocol (MCP) and FastAPI. We'll build a simple web application using FastAPI and convert it into an MCP server, enabling your L
 Dia-1.6B TTS : Best Text-to-Dialogue Generation Model - Analytics VidhyaMay 11, 2025 am 10:27 AM
Dia-1.6B TTS : Best Text-to-Dialogue Generation Model - Analytics VidhyaMay 11, 2025 am 10:27 AMExplore Dia-1.6B: A groundbreaking text-to-speech model developed by two undergraduates with zero funding! This 1.6 billion parameter model generates remarkably realistic speech, including nonverbal cues like laughter and sneezes. This article guide
 3 Ways AI Can Make Mentorship More Meaningful Than EverMay 10, 2025 am 11:17 AM
3 Ways AI Can Make Mentorship More Meaningful Than EverMay 10, 2025 am 11:17 AMI wholeheartedly agree. My success is inextricably linked to the guidance of my mentors. Their insights, particularly regarding business management, formed the bedrock of my beliefs and practices. This experience underscores my commitment to mentor
 AI Unearths New Potential In The Mining IndustryMay 10, 2025 am 11:16 AM
AI Unearths New Potential In The Mining IndustryMay 10, 2025 am 11:16 AMAI Enhanced Mining Equipment The mining operation environment is harsh and dangerous. Artificial intelligence systems help improve overall efficiency and security by removing humans from the most dangerous environments and enhancing human capabilities. Artificial intelligence is increasingly used to power autonomous trucks, drills and loaders used in mining operations. These AI-powered vehicles can operate accurately in hazardous environments, thereby increasing safety and productivity. Some companies have developed autonomous mining vehicles for large-scale mining operations. Equipment operating in challenging environments requires ongoing maintenance. However, maintenance can keep critical devices offline and consume resources. More precise maintenance means increased uptime for expensive and necessary equipment and significant cost savings. AI-driven
 Why AI Agents Will Trigger The Biggest Workplace Revolution In 25 YearsMay 10, 2025 am 11:15 AM
Why AI Agents Will Trigger The Biggest Workplace Revolution In 25 YearsMay 10, 2025 am 11:15 AMMarc Benioff, Salesforce CEO, predicts a monumental workplace revolution driven by AI agents, a transformation already underway within Salesforce and its client base. He envisions a shift from traditional markets to a vastly larger market focused on
 AI HR Is Going To Rock Our Worlds As AI Adoption SoarsMay 10, 2025 am 11:14 AM
AI HR Is Going To Rock Our Worlds As AI Adoption SoarsMay 10, 2025 am 11:14 AMThe Rise of AI in HR: Navigating a Workforce with Robot Colleagues The integration of AI into human resources (HR) is no longer a futuristic concept; it's rapidly becoming the new reality. This shift impacts both HR professionals and employees, dem


Hot AI Tools

Undresser.AI Undress
AI-powered app for creating realistic nude photos

AI Clothes Remover
Online AI tool for removing clothes from photos.

Undress AI Tool
Undress images for free

Clothoff.io
AI clothes remover

Video Face Swap
Swap faces in any video effortlessly with our completely free AI face swap tool!

Hot Article

Hot Tools

Notepad++7.3.1
Easy-to-use and free code editor

SublimeText3 Chinese version
Chinese version, very easy to use

Zend Studio 13.0.1
Powerful PHP integrated development environment

SublimeText3 Linux new version
SublimeText3 Linux latest version

WebStorm Mac version
Useful JavaScript development tools






